Development and Application of an Automated Raman Sensor for Bioprocess Monitoring: From the Laboratory to an Algae Production Platform
Abstract
:1. Introduction
2. Materials and Methods
2.1. Microalgal Strain and Culture Conditions
2.2. Determination of the Cell Density and the Concentration of Total Chlorophyll and Carotenoids
2.3. Lipid Extraction and Analysis
2.4. Determination of Nitrate Concentration
2.5. Development of the Raman Spectroscopy Measurement Approach
2.5.1. Experimental Set-Up
2.5.2. Raman Spectrometer and Parameters of Acquisition
2.6. Spectral Pre-Processing and Data Analysis
3. Results
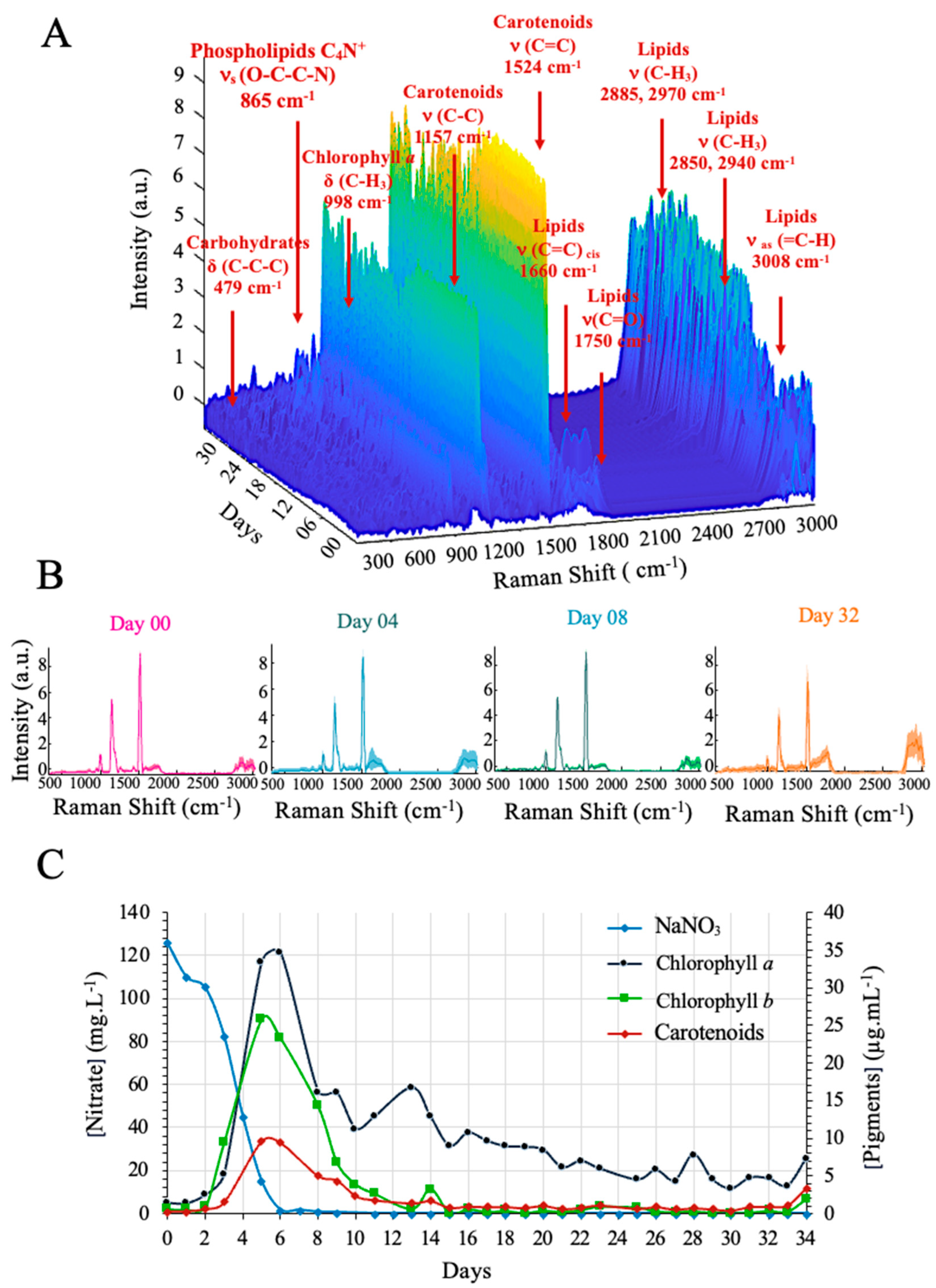
3.1. Microalgal Culture Monitoring at the Laboratory Scale
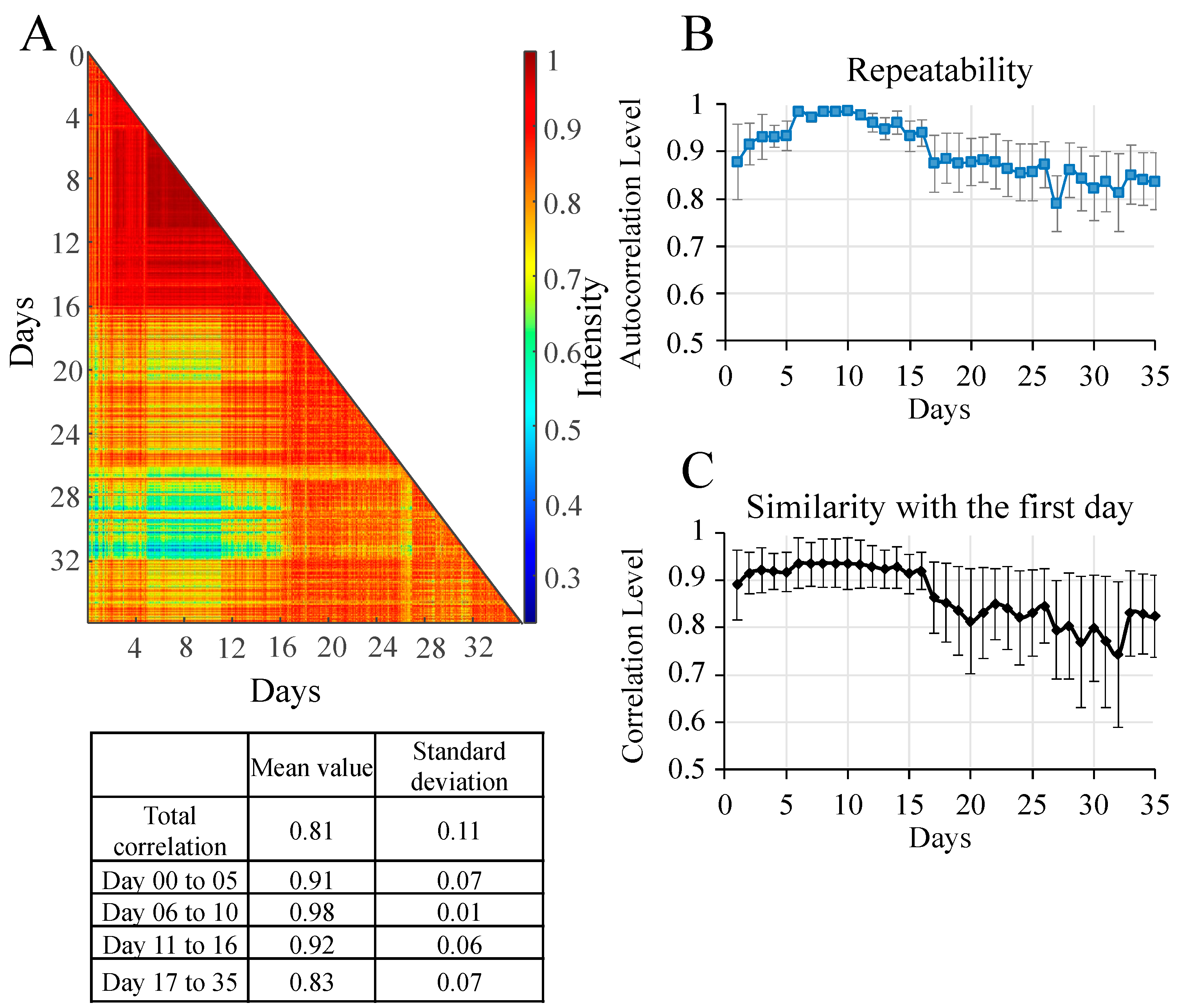
3.2. Verification of Repeatability and Similarity of Raman Measurements
3.3. Validation of the Developed Approach at the Pilot Scale
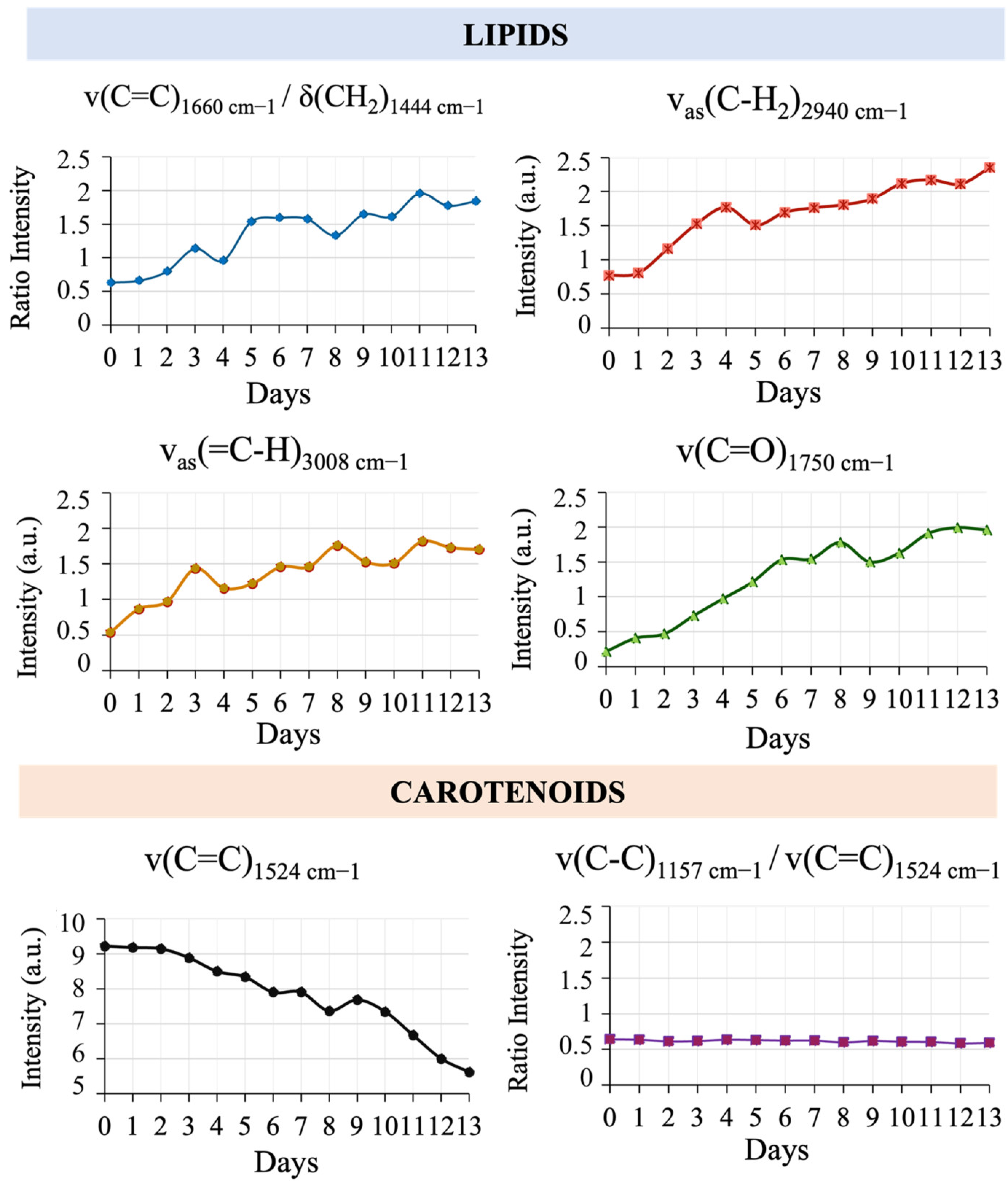
3.4. Chemometric Analysis of the Spectral Database
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jin, E.S.; Melis, A. Microalgal Biotechnology: Carotenoid Production by the Green algae Dunaliella salina. Biotechnol. Bioprocess Eng. 2003, 8, 331. [Google Scholar] [CrossRef]
- Grobbelaar, J.U. Algal Nutrition—Mineral Nutrition—Handbook of Microalgal Culture—Wiley Online Library. Available online: https://onlinelibrary.wiley.com/doi/abs/10.1002/9780470995280.ch6 (accessed on 21 April 2023).
- Wijffels, R.H.; Barbosa, M.J. An Outlook on Microalgal Biofuels. Science 2010, 329, 796–799. [Google Scholar] [CrossRef] [PubMed]
- Draaisma, R.B.; Wijffels, R.H.; Slegers, P.E.; Brentner, L.B.; Roy, A.; Barbosa, M.J. Food Commodities from Microalgae. Curr. Opin. Biotechnol. 2013, 24, 169–177. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.I.; Shin, J.H.; Kim, J.D. The Promising Future of Microalgae: Current Status, Challenges, and Optimization of a Sustainable and Renewable Industry for Biofuels, Feed, and Other Products. Microb. Cell Factories 2018, 17, 36. [Google Scholar] [CrossRef] [PubMed]
- Markou, G.; Nerantzis, E. Microalgae for High-Value Compounds and Biofuels Production: A Review with Focus on Cultivation under Stress Conditions. Biotechnol. Adv. 2013, 31, 1532–1542. [Google Scholar] [CrossRef]
- Vojinović, V.; Cabral, J.M.S.; Fonseca, L.P. Real-Time Bioprocess Monitoring: Part I: In Situ Sensors. Sens. Actuators B Chem. 2006, 114, 1083–1091. [Google Scholar] [CrossRef]
- Harms, P.; Kostov, Y.; Rao, G. Bioprocess Monitoring. Curr. Opin. Biotechnol. 2002, 13, 124–127. [Google Scholar] [CrossRef]
- Havlik, I.; Lindner, P.; Scheper, T.; Reardon, K.F. On-Line Monitoring of Large Cultivations of Microalgae and Cyanobacteria. Trends Biotechnol. 2013, 31, 406–414. [Google Scholar] [CrossRef]
- Elisabeth, B.; Filali, R.; Taidi, B. Microalgae Culture Quality Indicators: A Review. Crit. Rev. Biotechnol. 2021, 41, 457–473. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Grits, Y.A.; Etzion, D.; Ning, Z.; Richmond, A. Optical Properties of Nannochloropsis Sp and Their Application to Remote Estimation of Cell Mass. Biotechnol. Bioeng. 2000, 69, 516–525. [Google Scholar] [CrossRef]
- Esmonde-White, K.A.; Cuellar, M.; Uerpmann, C.; Lenain, B.; Lewis, I.R. Raman Spectroscopy as a Process Analytical Technology for Pharmaceutical Manufacturing and Bioprocessing. Anal. Bioanal. Chem. 2017, 409, 637–649. [Google Scholar] [CrossRef]
- Gómez-Caravaca, A.M.; Maggio, R.M.; Cerretani, L. Chemometric Applications to Assess Quality and Critical Parameters of Virgin and Extra-Virgin Olive Oil. A Review. Anal. Chim. Acta 2016, 913, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Karoui, R.; Downey, G.; Blecker, C. Mid-Infrared Spectroscopy Coupled with Chemometrics: A Tool for the Analysis of Intact Food Systems and the Exploration of Their Molecular Structure−Quality Relationships—A Review. Chem. Rev. 2010, 110, 6144–6168. [Google Scholar] [CrossRef]
- Adar, F.; Geiger, R.; Noonan, J. Raman Spectroscopy for Process/Quality Control. Appl. Spectrosc. Rev. 1997, 32, 45–101. [Google Scholar] [CrossRef]
- Rostron, P.; Gerber, D. Raman Spectroscopy, a Review. Int. J. Eng. Tech. Res. 2016, 6, 50–64. [Google Scholar]
- Parab, N.; Tomar, V. Raman Spectroscopy of Algae: A Review. J. Nanomedic Nanotechnol. 2012, 3, 24. [Google Scholar] [CrossRef]
- Huang, Y.Y.; Beal, C.M.; Cai, W.W.; Ruoff, R.S.; Terentjev, E.M. Micro-Raman Spectroscopy of Algae: Composition Analysis and Fluorescence Background Behavior. Biotechnol. Bioeng. 2010, 105, 889–898. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.K.; Nelson, D.R.; Abdrabu, R.; Khraiwesh, B.; Jijakli, K.; Arnoux, M.; O’Connor, M.J.; Bahmani, T.; Cai, H.; Khapli, S.; et al. An Integrative Raman Microscopy-Based Workflow for Rapid in Situ Analysis of Microalgal Lipid Bodies. Biotechnol. Biofuels 2015, 8, 164. [Google Scholar] [CrossRef]
- Shao, Y.; Gu, W.; Jiang, L.; Zhu, Y.; Gong, A. Study on the Visualization of Pigment in Haematococcus pluvialis by Raman Spectroscopy Technique. Sci. Rep. 2019, 9, 12097. [Google Scholar] [CrossRef]
- Samek, O.; Zemanek, P.; Jonas, A.; Telle, H. Characterization of Oil-producing Microalgae Using Raman Spectroscopy. Laser Phys. Lett. 2011, 8, 701–709. [Google Scholar] [CrossRef]
- Brahma, S.K.; Hargraves, P.E.; Howard, W.F.; Nelson, W.H. A Resonance Raman Method for the Rapid Detection and Identification of Algae in Water. Appl. Spectrosc. 1983, 37, 55–58. [Google Scholar] [CrossRef]
- Howard, W.F.; Nelson, W.H.; Sperry, J.F. A Resonance Raman Method for the Rapid Detection and Identification of Bacteria in Water. Appl. Spectrosc. 1980, 34, 72–75. [Google Scholar] [CrossRef]
- Wu, Q.; Nelson, W.H.; Hargraves, P.; Zhang, J.; Brown, C.W.; Seelenbinder, J.A. Differentiation of Algae Clones on the Basis of Resonance Raman Spectra Excited by Visible Light. Anal. Chem. 1998, 70, 1782–1787. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Fang, S.; Xie, W.; Zhang, P.; Li, Z.; Zhou, D.; Zhang, Z.; Guo, J.; Du, C.; Du, J.; et al. Assessment of Physiological Responses and Growth Phases of Different Microalgae under Environmental Changes by Raman Spectroscopy with Chemometrics. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2018, 204, 287–294. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Liu, X.; Fang, H.; Zhang, J.; Feng, X. Real-Time and In Situ Evaluation of Phycocyanin Concentration in Spirulina Platensis Cultivation System by Using Portable Raman Spectroscopy. J. Chem. 2021, 2021, e8857984. [Google Scholar] [CrossRef]
- Lieutaud, C.; Assaf, A.; Gonçalves, O.; Wielgosz-Collin, G.; Thouand, G. Fast Non-Invasive Monitoring of Microalgal Physiological Stage in Photobioreactors through Raman Spectroscopy. Algal Res. 2019, 42, 101595. [Google Scholar] [CrossRef]
- Moudříková, Š.; Ivanov, I.N.; Vítová, M.; Nedbal, L.; Zachleder, V.; Mojzeš, P.; Bišová, K. Comparing Biochemical and Raman Microscopy Analyses of Starch, Lipids, Polyphosphate, and Guanine Pools during the Cell Cycle of Desmodesmus quadricauda. Cells 2021, 10, 62. [Google Scholar] [CrossRef]
- Wang, T.; Ji, Y.; Wang, Y.; Jia, J.; Li, J.; Huang, S.; Han, D.; Hu, Q.; Huang, W.E.; Xu, J. Quantitative Dynamics of Triacylglycerol Accumulation in Microalgae Populations at Single-Cell Resolution Revealed by Raman Microspectroscopy. Biotechnol. Biofuels 2014, 7, 58. [Google Scholar] [CrossRef]
- Fagerer, S.R.; Schmid, T.; Ibáñez, A.J.; Pabst, M.; Steinhoff, R.; Jefimovs, K.; Urban, P.L.; Zenobi, R. Analysis of Single Algal Cells by Combining Mass Spectrometry with Raman and Fluorescence Mapping. Analyst 2013, 138, 6732–6736. [Google Scholar] [CrossRef]
- Zhang, P.; Xiao, Y.; Li, Z.; Guo, J.; Lu, L. Microalgae in Microwell Arrays Exhibit Differences with Those in Flasks: Evidence from Growth Rate, Cellular Carotenoid, and Oxygen Production. Front. Plant Sci. 2018, 8, 2251. [Google Scholar] [CrossRef]
- Xie, P.; Chen, C.; Zhang, C.; Su, G.; Ren, N.; Ho, S.-H. Revealing the Role of Adsorption in Ciprofloxacin and Sulfadiazine Elimination Routes in Microalgae. Water Res. 2020, 172, 115475. [Google Scholar] [CrossRef] [PubMed]
- Nadadoor, V.R.; De la Hoz Siegler, H.; Shah, S.L.; McCaffrey, W.C.; Ben-Zvi, A. Online Sensor for Monitoring a Microalgal Bioreactor System Using Support Vector Regression. Chemom. Intell. Lab. Syst. 2012, 110, 38–48. [Google Scholar] [CrossRef]
- Noack, K.; Eskofier, B.; Kiefer, J.; Dilk, C.; Bilow, G.; Schirmer, M.; Buchholz, R.; Leipertz, A. Combined shifted-excitation Raman difference spectroscopy and support vector regression for monitoring the algal production of complex polysaccharides. Analyst 2013, 138, 5639–5646. [Google Scholar] [CrossRef] [PubMed]
- Podevin, M.; Fotidis, I.A.; Angelidaki, I. Microalgal Process-Monitoring Based on High-Selectivity Spectroscopy Tools: Status and Future Perspectives. Crit. Rev. Biotechnol. 2018, 38, 704–718. [Google Scholar] [CrossRef] [PubMed]
- Wood, B.R.; Heraud, P.; Stojkovic, S.; Morrison, D.; Beardall, J.; McNaughton, D. A Portable Raman Acoustic Levitation Spectroscopic System for the Identification and Environmental Monitoring of Algal Cells. Anal. Chem. 2005, 77, 4955–4961. [Google Scholar] [CrossRef] [PubMed]
- Davis, R.W.; Wu, H.; Singh, S. Multispectral Sorter for Rapid, Nondestructive Optical Bioprospecting for Algae Biofuels. In Proceedings of the Imaging, Manipulation, and Analysis of Biomolecules, Cells, and Tissues XII, SPIE, San Francisco, CA, USA, 4 March 2014; Volume 8947, pp. 252–262. [Google Scholar]
- Pilát, Z.; Ježeka, J.; KaÅka, J.; Zemánek, P. Raman Tweezers in Microfluidic Systems for Analysis and Sorting of Living Cells. In Proceedings of the Imaging, Manipulation, and Analysis of Biomolecules, Cells, and Tissues XII, SPIE, San Francisco, CA, USA, 4 March 2014; Volume 9441. [Google Scholar]
- Mattila-Sandholm, T.; Wirtanen, G. Biofilm Formation in the Industry: A Review. Food Rev. Int. 1992, 8, 573–603. [Google Scholar] [CrossRef]
- Bezerra, M.A.; Lemos, V.A.; de Oliveira, D.M.; Novaes, C.G.; Barreto, J.A.; Alves, J.P.S.; da Mata Cerqueira, U.M.F.; Santos, Q.O.d.; Araújo, S.A. Automation of Continuous Flow Analysis Systems—A Review. Microchem. J. 2020, 155, 104731. [Google Scholar] [CrossRef]
- Piasecka, A.; Cieśla, J.; Koczańska, M.; Krzemińska, I. Effectiveness of Parachlorella Kessleri Cell Disruption Evaluated with the Use of Laser Light Scattering Methods. J. Appl. Phycol. 2019, 31, 97–107. [Google Scholar] [CrossRef]
- Krzemińska, I.; Nawrocka, A.; Piasecka, A.; Jagielski, P.; Tys, J. Cultivation of Chlorella Protothecoides in Photobioreactors: The Combined Impact of Photoperiod and CO2 Concentration. Eng. Life Sci. 2015, 15, 533–541. [Google Scholar] [CrossRef]
- Ritchie, R.J. Consistent Sets of Spectrophotometric Chlorophyll Equations for Acetone, Methanol and Ethanol Solvents. Photosynth. Res. 2006, 89, 27–41. [Google Scholar] [CrossRef]
- Strickland, J.D.H.; Parsons, T.R. A Practical Handbook of Seawater Analysis, 2nd ed.; Fisheries Research Board of Canada: Ottawa, ON, Canada, 1972. [Google Scholar] [CrossRef]
- Bligh, E.G.; Dyer, W.J. A Rapid Method of Total Lipid Extraction and Purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef]
- Gross, M.; Zhao, X.; Mascarenhas, V.; Wen, Z. Effects of the Surface Physico-Chemical Properties and the Surface Textures on the Initial Colonization and the Attached Growth in Algal Biofilm. Biotechnol. Biofuels 2016, 9, 38. [Google Scholar] [CrossRef]
- Fiche Générale Relative à la Réglementation des Matériaux au Contact des Denrées Alimentaires. Available online: https://www.economie.gouv.fr/dgccrf/Fiche-generale-relative-a-la-reglementation-des-ma (accessed on 11 April 2023).
- Lieutaud, C. Mesure En Ligne Par Spectroscopie Raman Appliquée à Un Photobioréacteur de Microalgues. Ph.D. Thesis, Nantes Universit, Nantes, France, 2019. [Google Scholar]
- Cordella, C.B.Y.; Bertrand, D. SAISIR: A New General Chemometric Toolbox. TrAC Trends Anal. Chem. 2014, 54, 75–82. [Google Scholar] [CrossRef]
- Abdi, H.; Williams, L.J. Principal Component Analysis. WIREs Comput. Stat. 2010, 2, 433–459. [Google Scholar] [CrossRef]
- Saito, S.; Tasumi, M. Normal-Coordinate Analysis of β-Carotene Isomers and Assignments of the Raman and Infrared Bands. J. Raman Spectrosc. 1983, 14, 310–321. [Google Scholar] [CrossRef]
- Lee, T.-H.; Chang, J.-S.; Wang, H.-Y. Rapid and in Vivo Quantification of Cellular Lipids in Chlorella vulgaris Using Near-Infrared Raman Spectrometry. Anal. Chem. 2013, 85, 2155–2160. [Google Scholar] [CrossRef]
- Schulz, H.; Baranska, M. Identification and Quantification of Valuable Plant Substances by IR and Raman Spectroscopy. Vib. Spectrosc. 2007, 43, 13–25. [Google Scholar] [CrossRef]
- Heraud, P.; Beardall, J.; McNaughton, D.; Wood, B.R. In Vivo Prediction of the Nutrient Status of Individual Microalgal Cells Using Raman Microspectroscopy. FEMS Microbiol. Lett. 2007, 275, 24–30. [Google Scholar] [CrossRef] [PubMed]
- Procházková, G.; Brányiková, I.; Zachleder, V.; Brányik, T. Effect of Nutrient Supply Status on Biomass Composition of Eukaryotic Green Microalgae. J. Appl. Phycol. 2014, 26, 1359–1377. [Google Scholar] [CrossRef]
- Zachleder, V.; Ivanov, I.N.; Kselíková, V.; Bialevich, V.; Vítová, M.; Ota, S.; Takeshita, T.; Kawano, S.; Bišová, K. Characterization of Growth and Cell Cycle Events Affected by Light Intensity in the Green Alga Parachlorella kessleri: A New Model for Cell Cycle Research. Biomolecules 2021, 11, 891. [Google Scholar] [CrossRef]
- Kandilian, R.; Taleb, A.; Heredia, V.; Cogne, G.; Pruvost, J. Effect of Light Absorption Rate and Nitrate Concentration on TAG Accumulation and Productivity of Parachlorella kessleri Cultures Grown in Chemostat Mode. Algal Res. Biomass Biofuels Bioprod. 2019, 39, 101442. [Google Scholar] [CrossRef]
- Brányiková, I.; Maršálková, B.; Doucha, J.; Brányik, T.; Bišová, K.; Zachleder, V.; Vítová, M. Microalgae—Novel Highly Efficient Starch Producers. Biotechnol. Bioeng. 2011, 108, 766–776. [Google Scholar] [CrossRef] [PubMed]
- Clavijo Rivera, E.; Montalescot, V.; Viau, M.; Drouin, D.; Bourseau, P.; Frappart, M.; Monteux, C.; Couallier, E. Mechanical Cell Disruption of Parachlorella kessleri Microalgae: Impact on Lipid Fraction Composition. Bioresour. Technol. 2018, 256, 77–85. [Google Scholar] [CrossRef]
- Wu, H.; Volponi, J.V.; Oliver, A.E.; Parikh, A.N.; Simmons, B.A.; Singh, S. In Vivo Lipidomics Using Single-Cell Raman Spectroscopy. Proc. Natl. Acad. Sci. USA 2011, 108, 3809–3814. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, V.E.; Castro, H.V.; Edwards, H.G.M.; de Oliveira, L.F.C. Carotenes and Carotenoids in Natural Biological Samples: A Raman Spectroscopic Analysis. J. Raman Spectrosc. 2010, 41, 642–650. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wieser, W.; Assaf, A.A.; Le Gouic, B.; Dechandol, E.; Herve, L.; Louineau, T.; Dib, O.H.; Gonçalves, O.; Titica, M.; Couzinet-Mossion, A.; et al. Development and Application of an Automated Raman Sensor for Bioprocess Monitoring: From the Laboratory to an Algae Production Platform. Sensors 2023, 23, 9746. https://doi.org/10.3390/s23249746
Wieser W, Assaf AA, Le Gouic B, Dechandol E, Herve L, Louineau T, Dib OH, Gonçalves O, Titica M, Couzinet-Mossion A, et al. Development and Application of an Automated Raman Sensor for Bioprocess Monitoring: From the Laboratory to an Algae Production Platform. Sensors. 2023; 23(24):9746. https://doi.org/10.3390/s23249746
Chicago/Turabian StyleWieser, Wiviane, Antony Ali Assaf, Benjamin Le Gouic, Emmanuel Dechandol, Laura Herve, Thomas Louineau, Omar Hussein Dib, Olivier Gonçalves, Mariana Titica, Aurélie Couzinet-Mossion, and et al. 2023. "Development and Application of an Automated Raman Sensor for Bioprocess Monitoring: From the Laboratory to an Algae Production Platform" Sensors 23, no. 24: 9746. https://doi.org/10.3390/s23249746






