LeafSpec-Dicot: An Accurate and Portable Hyperspectral Imaging Device for Dicot Leaves
Abstract
:Highlights
- The first portable hyperspectral imaging device specially designed for dicot plants to capture the image of an entire soybean leaf.
- The prediction of nitrogen content using images captured from the device establishes a strong correlation with the nitrogen content measured via chemical analysis.
- The imaging process is fully automated to maintain the consistency of images and relive the labors from operators.
- The device allows users to see the leaf more clearly which could open new pathways for plant study.
Abstract
1. Introduction
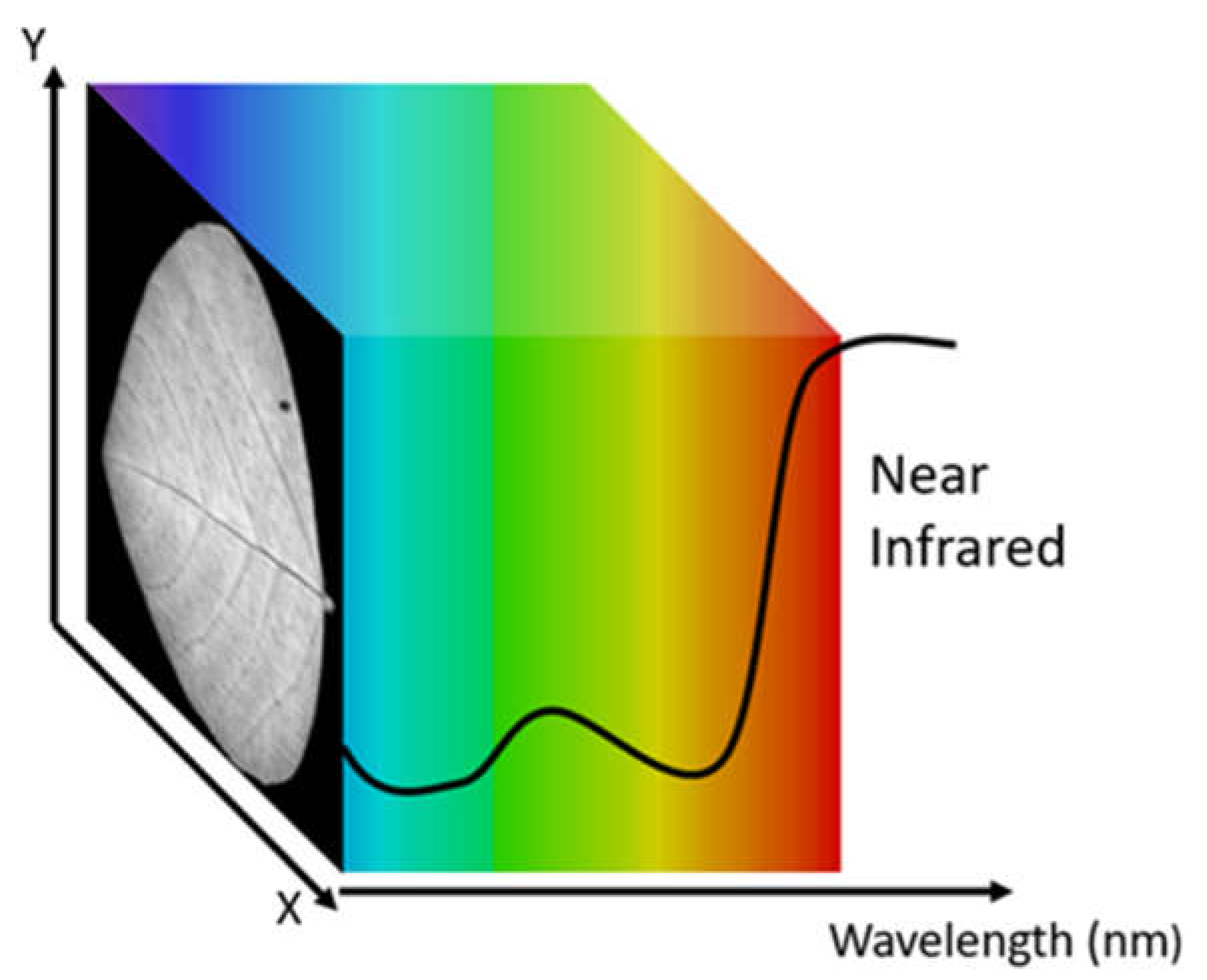
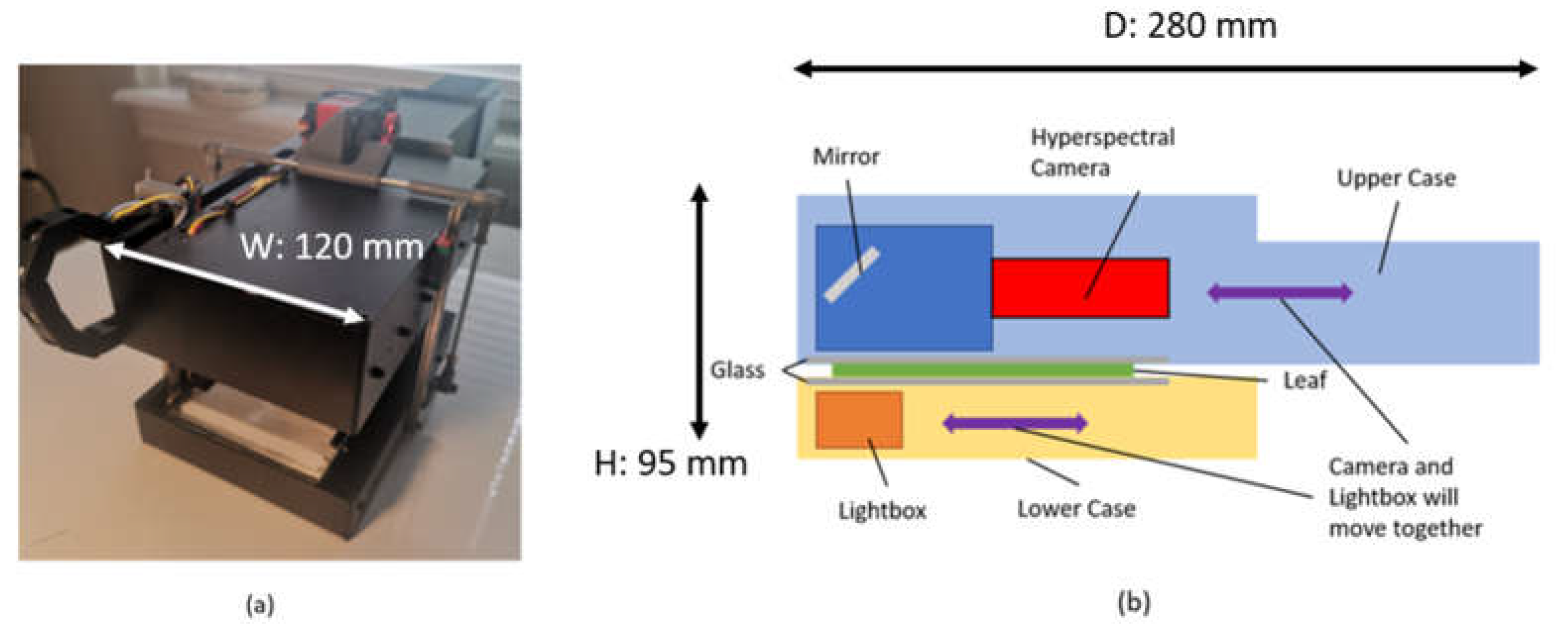
2. Hardware Development
2.1. Overview
2.2. Hardware Design
2.2.1. Lightbox
2.2.2. Scanning Mechanism
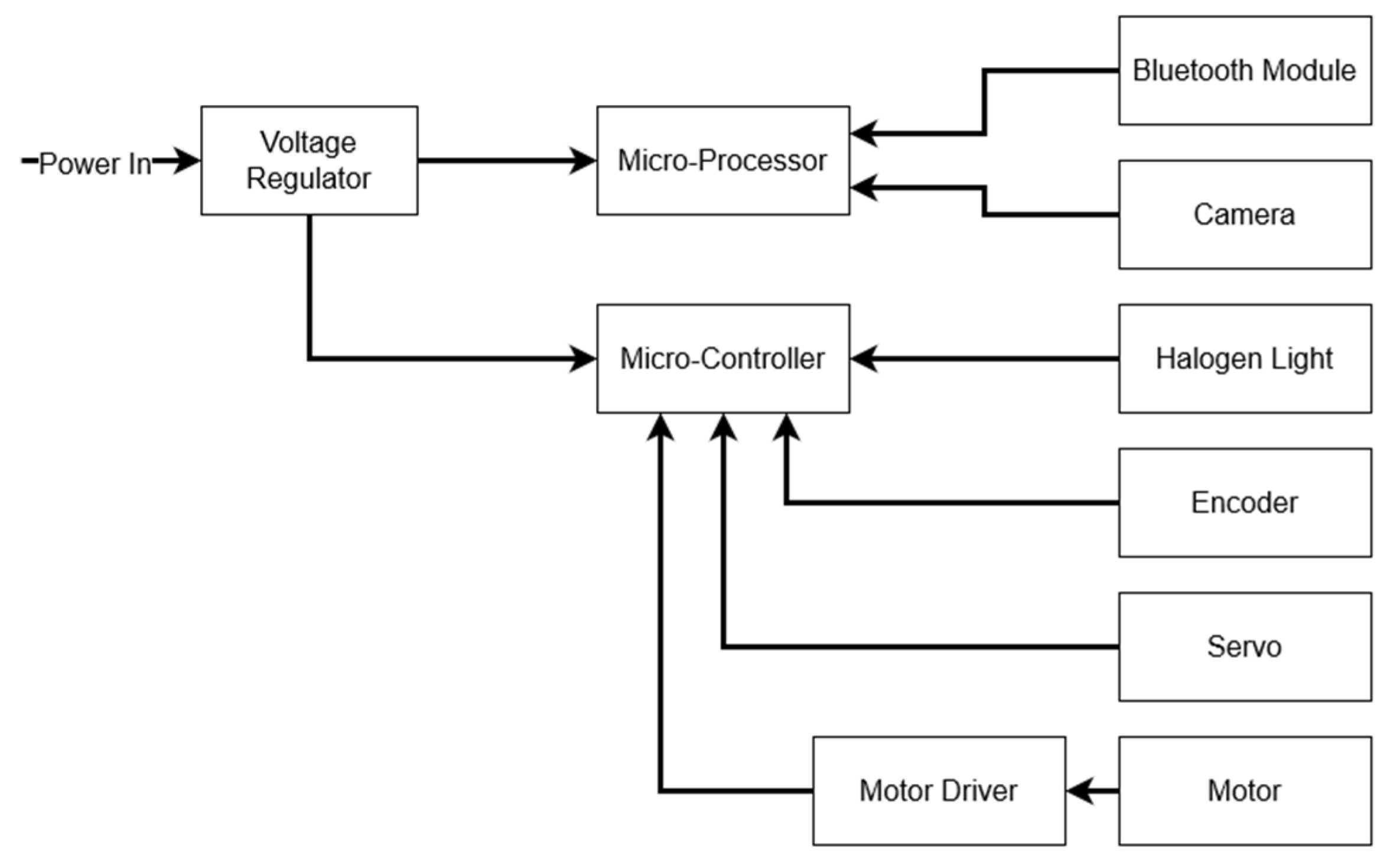
2.2.3. Electronics
Power Supply
Microprocessor
Microcontroller
2.3. Device Operation and Data Flow
3. Validation of the Effectiveness of the Device through Correlating NDVI with Nitrogen Content of Soybean Plants
3.1. Overview
3.2. Data Collection
3.2.1. Experimental Setup
3.2.2. Collection of HS Images
3.2.3. Laboratory-Tested Nutrient Data Collection
3.3. Data Analysis
3.3.1. Pre-Processing and Modeling Setup
3.3.2. Modeling Using Mean NDVI Method
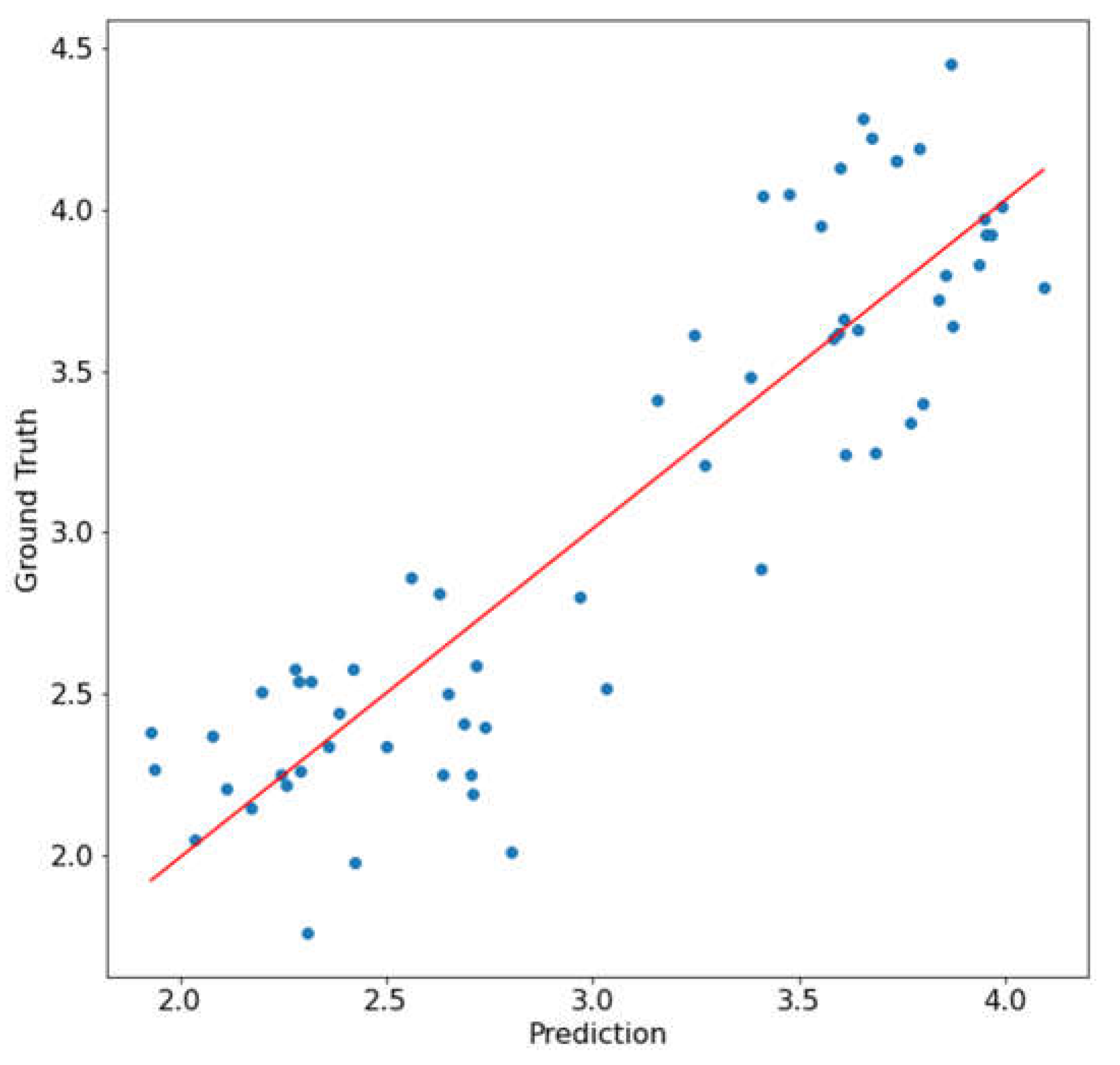
3.3.3. Mean NDVI Method Correlation Result
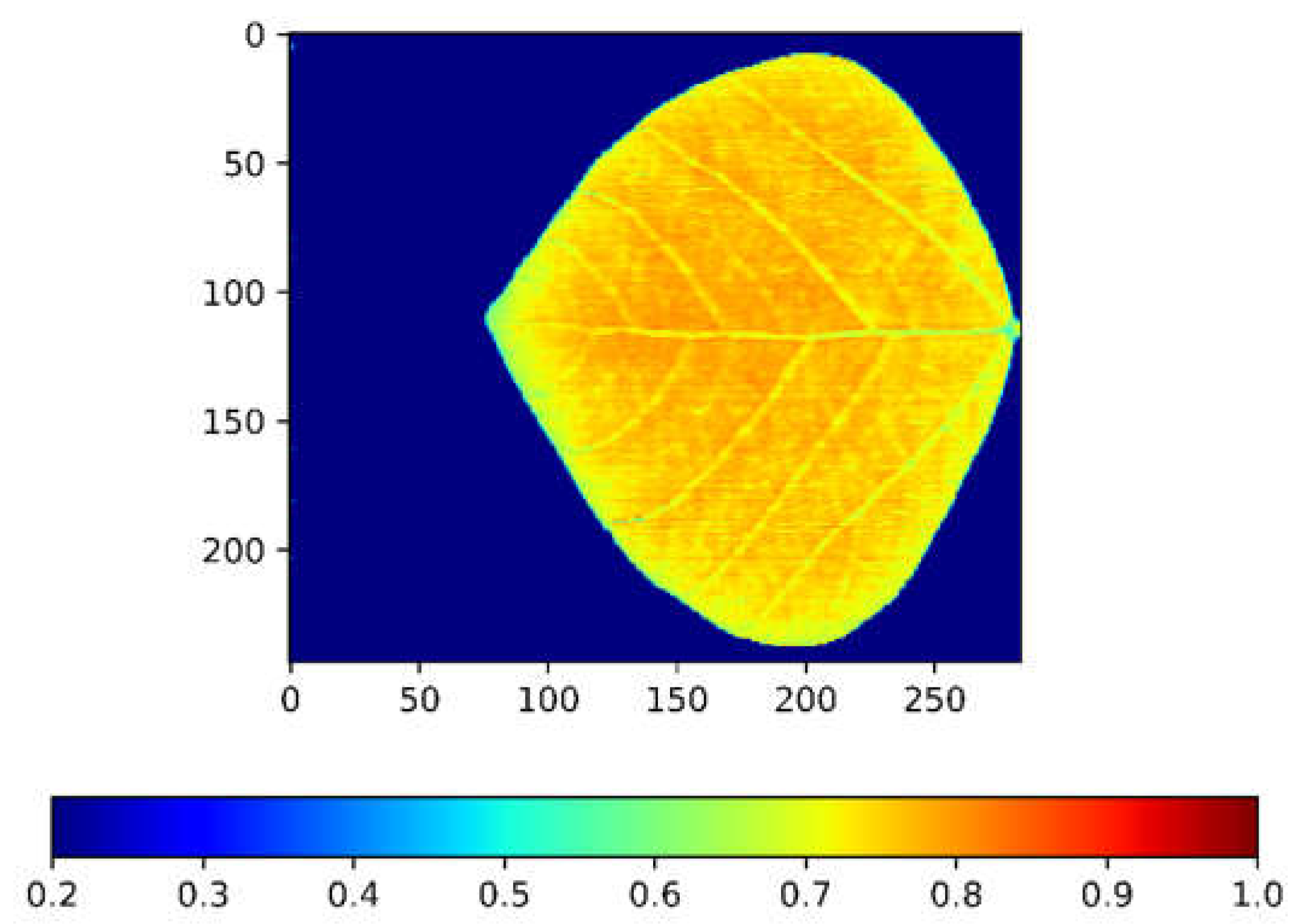
3.3.4. Modeling Using Whole-Leaf NDVI Heatmap
3.3.5. Whole-Leaf NDVI Heatmap Correlation Result
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Li, L.; Zhang, Q.; Huang, D. A review of imaging techniques for plant phenotyping. Sensors 2014, 14, 20078–20111. [Google Scholar] [CrossRef] [PubMed]
- Godfray, H.C.J.; Beddington, J.R.; Crute, I.R.; Haddad, L.; Lawrence, D.; Muir, J.F.; Pretty, J.; Robinson, S.; Thomas, S.M.; Toulmin, C. Food Security: The Challenge of Feeding 9 Billion People. Science 2010, 327, 812–818. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Masuda, T.; Goldsmith, P.D. World soybean production: Area harvested, yield, and long-term projections. Int. Food Agribus. Manag. Rev. 2009, 12, 143–162. [Google Scholar] [CrossRef]
- Pagano, M.C.; Miransari, M. The importance of soybean production worldwide. In Abiotic and Biotic Stresses in Soybean Production; Elsevier Inc.: Amsterdam, The Netherlands, 2016. [Google Scholar] [CrossRef]
- Hagely, K.; Konda, A.R.; Kim, J.-H.; Cahoon, E.B.; Bilyeu, K. Molecular-assisted breeding for soybean with high oleic/low linolenic acid and elevated vitamin E in the seed oil. Mol. Breed. 2021, 41, 3. [Google Scholar] [CrossRef]
- Gowen, A.A.; O’Donnell, C.P.; Cullen, P.J.; Downey, G.; Frias, J.M. Hyperspectral imaging—An emerging process analytical tool for food quality and safety control. Trends Food Sci. Technol. 2007, 18, 590–598. [Google Scholar] [CrossRef]
- Ge, Y.; Bai, G.; Stoerger, V.; Schnable, J.C. Temporal dynamics of maize plant growth, water use, and leaf water content using automated high throughput RGB and hyperspectral imaging. Comput. Electron. Agric. 2016, 127, 625–632. [Google Scholar] [CrossRef] [Green Version]
- Li, D.; Quan, C.; Song, Z.; Li, X.; Yu, G.; Li, C.; Muhammad, A. High-Throughput Plant Phenotyping Platform (HT3P) as a Novel Tool for Estimating Agronomic Traits from the Lab to the Field. Front. Bioeng. Biotechnol. 2021, 8, 623705. [Google Scholar] [CrossRef]
- Chen, D.; Neumann, K.; Friedel, S.; Kilian, B.; Chen, M.; Altmann, T.; Klukas, C. Dissecting the Phenotypic Components of Crop Plant Growth and Drought Responses Based on High-Throughput Image Analysis. Plant Cell 2014, 26, 4636–4655. [Google Scholar] [CrossRef] [Green Version]
- Ma, D.; Carpenter, N.; Amatya, S.; Maki, H.; Wang, L.; Zhang, L.; Neeno, S.; Tuinstra, M.R.; Jin, J. Removal of greenhouse microclimate heterogeneity with conveyor system for indoor phenotyping. Comput. Electron. Agric. 2019, 166, 104979. [Google Scholar] [CrossRef]
- Miao, C.; Guo, A.; Thompson, A.M.; Yang, J.; Ge, Y.; Schnable, J.C. Automation of leaf counting in maize and sorghum using deep learning. Plant Phenome J. 2021, 4, e20022. [Google Scholar] [CrossRef]
- Miao, C.; Xu, Y.; Liu, S.; Schnable, P.S.; Schnable, J.C. Increased Power and Accuracy of Causal Locus Identification in Time Series Genome-wide Association in Sorghum. Plant Physiol. 2020, 183, 1898–1909. [Google Scholar] [CrossRef] [PubMed]
- Virlet, N.; Sabermanesh, K.; Sadeghi-Tehran, P.; Hawkesford, M.J. Field Scanalyzer: An automated robotic field phenotyping platform for detailed crop monitoring. Funct. Plant Biol. 2017, 44, 143–153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomas, S.; Behmann, J.; Steier, A.; Kraska, T.; Muller, O.; Rascher, U.; Mahlein, A.-K. Quantitative assessment of disease severity and rating of barley cultivars based on hyperspectral imaging in a non-invasive, automated phenotyping platform. Plant Methods 2018, 14, 45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lavado, R.S.; Porcelli, C.A.; Alvarez, R. Nutrient and heavy metal concentration and distribution in corn, soybean and wheat as affected by different tillage systems in the Argentine Pampas. Soil Tillage Res. 2001, 62, 55–60. [Google Scholar] [CrossRef]
- Yuan, Z.; Cao, Q.; Zhang, K.; Ata-Ul-Karim, S.T.; Tian, Y.; Zhu, Y.; Cao, W.; Liu, X. Optimal Leaf Positions for SPAD Meter Measurement in Rice. Front. Plant Sci. 2016, 7, 719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, J.; Li, C.; Wen, Y.; Gao, X.; Shi, F.; Han, L. Spatial distribution of SPAD value and determination of the suitable leaf for N diagnosis in cucumber. IOP Conf. Ser. Earth Environ. Sci. 2018, 108, 22001. [Google Scholar] [CrossRef]
- Vijayarangan, S.; Sodhi, P.; Kini, P.; Bourne, J.; Du, S.; Sun, H.; Poczos, B.; Apostolopoulos, D.; Wettergreen, D. High-Throughput Robotic Phenotyping of Energy Sorghum Crops. In Field and Service Robotics; Hutter, M., Siegwart, R., Eds.; Springer International Publishing: New York, NY, USA, 2018; pp. 99–113. [Google Scholar]
- Pölönen, I.; Saari, H.; Kaivosoja, J.; Honkavaara, E.; Pesonen, L. Hyperspectral imaging based biomass and nitrogen content estimations from light-weight UAV. Remote Sens. Agric. Ecosyst. Hydrolo. XV 2013, 8887, 88870J. [Google Scholar] [CrossRef]
- Alabi, T.R.; Abebe, A.T.; Chigeza, G.; Fowobaje, K.R. Estimation of soybean grain yield from multispectral high-resolution UAV data with machine learning models in West Africa. Remote Sens. Appl. Soc. Environ. 2022, 27, 100782. [Google Scholar] [CrossRef]
- Gano, B.; Dembele, J.S.B.; Ndour, A.; Luquet, D.; Beurier, G.; Diouf, D.; Audebert, A. Using UAV Borne, Multi-Spectral Imaging for the Field Phenotyping of Shoot Biomass, Leaf Area Index and Height of West African Sorghum Varieties under Two Contrasted Water Conditions. Agronomy 2021, 11, 850. [Google Scholar] [CrossRef]
- Maimaitijiang, M.; Ghulam, A.; Sidike, P.; Hartling, S.; Maimaitiyiming, M.; Peterson, K.; Shavers, E.; Fishman, J.; Peterson, J.; Kadam, S.; et al. Unmanned Aerial System (UAS)-based phenotyping of soybean using multi-sensor data fusion and extreme learning machine. ISPRS J. Photogramm. Remote Sens. 2017, 134, 43–58. [Google Scholar] [CrossRef]
- Bai, G.; Ge, Y.; Hussain, W.; Baenziger, P.S.; Graef, G. A multi-sensor system for high throughput field phenotyping in soybean and wheat breeding. Comput. Electron. Agric. 2016, 128, 181–192. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Y.; Li, C.; Robertson, J.S.; Sun, S.; Xu, R.; Paterson, A.H. GPhenoVision: A Ground Mobile System with Multi-modal Imaging for Field-Based High Throughput Phenotyping of Cotton. Sci. Rep. 2018, 8, 1213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deery, D.; Jimenez-Berni, J.; Jones, H.; Sirault, X.; Furbank, R. Proximal Remote Sensing Buggies and Potential Applications for Field-Based Phenotyping. Agronomy 2014, 4, 349–379. [Google Scholar] [CrossRef] [Green Version]
- Wendel, A.; Underwood, J.; Walsh, K. Maturity estimation of mangoes using hyperspectral imaging from a ground based mobile platform. Comput. Electron. Agric. 2018, 155, 298–313. [Google Scholar] [CrossRef]
- NASA. Incoming Sunlight. 2009. Available online: https://earthobservatory.nasa.gov/features/EnergyBalance/page2.php. (accessed on 10 December 2022).
- Ma, D.; Rehman, T.U.; Zhang, L.; Maki, H.; Tuinstra, M.R.; Jin, J. Modeling of diurnal changing patterns in airborne crop remote sensing images. Remote Sens. 2021, 13, 1719. [Google Scholar] [CrossRef]
- Kanning, M.; Kühling, I.; Trautz, D.; Jarmer, T. High-resolution UAV-based hyperspectral imagery for LAI and chlorophyll estimations from wheat for yield prediction. Remote Sens. 2018, 10, 2000. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Jin, J.; Song, Z.; Wang, J.; Zhang, L.; Rehman, T.U.; Ma, D.; Carpenter, N.R.; Tuinstra, M.R. LeafSpec: An accurate and portable hyperspectral corn leaf imager. Comput. Electron. Agric. 2020, 169, 105209. [Google Scholar] [CrossRef]
- Mishra, P.; Asaari, M.S.M.; Herrero-Langreo, A.; Lohumi, S.; Diezma, B.; Scheunders, P. Close range hyperspectral imaging of plants: A review. Biosyst. Eng. 2017, 164, 49–67. [Google Scholar] [CrossRef]
- Chen, Z.; Wang, J.; Wang, T.; Song, Z.; Li, Y.; Huang, Y.; Wang, L.; Jin, J. Automated in-field leaf-level hyperspectral imaging of corn plants using a Cartesian robotic platform. Comput. Electron. Agric. 2021, 183, 105996. [Google Scholar] [CrossRef]
- Cabrera-Bosquet, L.; Molero, G.; Stellacci, A.; Bort, J.; Nogués, S.; Araus, J. NDVI as a potential tool for predicting biomass, plant nitrogen content and growth in wheat genotypes subjected to different water and nitrogen conditions. Cereal Res. Commun. 2011, 39, 147–159. [Google Scholar] [CrossRef]
- Edalat, M.; Naderi, R.; Egan, T.P. Corn nitrogen management using NDVI and SPAD sensor-based data under conventional vs. reduced tillage systems. J. Plant Nutr. 2019, 42, 2310–2322. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. arXiv 2016, arXiv:1512.03385. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Chen, Z.; Wang, J.; Jin, J. LeafSpec-Dicot: An Accurate and Portable Hyperspectral Imaging Device for Dicot Leaves. Sensors 2023, 23, 3687. https://doi.org/10.3390/s23073687
Li X, Chen Z, Wang J, Jin J. LeafSpec-Dicot: An Accurate and Portable Hyperspectral Imaging Device for Dicot Leaves. Sensors. 2023; 23(7):3687. https://doi.org/10.3390/s23073687
Chicago/Turabian StyleLi, Xuan, Ziling Chen, Jialei Wang, and Jian Jin. 2023. "LeafSpec-Dicot: An Accurate and Portable Hyperspectral Imaging Device for Dicot Leaves" Sensors 23, no. 7: 3687. https://doi.org/10.3390/s23073687




