Bioavailability of Cd, Zn and Hg in Soil to Nine Recombinant Luminescent Metal Sensor Bacteria
Abstract
:1. Introduction
- To investigate the effect of host bacterium (Gram-positive or -negative), genetic metal-response element and location of the metal-response element (plasmid or chromosome) on sensitivity (limit of determination and toxicity) of sensor bacteria towards target heavy metals
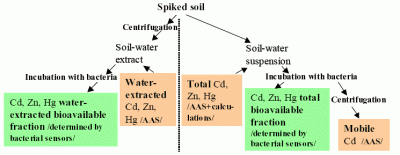
- To evaluate total bioavailable and water-extracted bioavailable fractions of Cd, Zn and Hg in spiked soils using sensor bacteria belonging to both Gram-negative (Pseudomonas fluorescens and Escherichia coli) as well as Gram-positive (Bacillus subtilis and Staphylococcus aureus) bacterial families thus representing organisms of different natural habitats, physiology and cell wall structure.
- To compare the total and water-extracted bioavailable fractions measured by different recombinant bacterial sensors in order to investigate whether the bioavailability of metals depends on the type of bacterial cell or nature of the metal-response element used for the construction of the sensors.
- Using Cd as a model, to monitor the changes in bioavailability of Cd as a result of bacterial metabolic activity during 2-hour incubation of sensor bacteria with soil-water extracts and suspensions.
2. Results and Discussion
2.1. Characterization of bacterial sensors
2.2. Response of the sensor strains to Hg, Cd and Zn
2.3. Bioavailability of Cd, Zn and Hg in soil
2.4. Differences in bioavailability of Cd, Zn and Hg in soil to different sensor bacteria
2.5. Time-dependent changes in bioavailability of Cd in soil
3. Materials and Methods
3.1 Bacterial strains
3.2. Cultivation of bacteria
3.3. Soil samples and their preparation
3.4. Bioavailability tests and calculations
3.5. Fluorescence microscopy
3.6. Measurement of mobile Cd from soil
Conclusions
- The limit of determination of the sensors was determined mainly by the type of the genetic metal-response element used for the construction of the sensor bacteria. At the same time, toxicity of the Cd, Zn and Hg standard solutions was mostly dependent on the host bacterium, Gram-positive bacteria being in general more sensitive to all the metals than Gram-negative.
- Bioavailability of Cd, Zn and Hg in soil did not depend neither on the limit of determination (determined according to standard calibration curve) of the used sensor nor on the metal-response elements expressed in these sensor cells.
- The water-extracted bioavailable fractions of Zn, Cd and Hg were low (making 0.24 – 0.37%, 0.19 – 0.46 % and 1.7 – 4.9 % of the total Zn Cd and Hg, respectively) and similar to all the used sensor strains.
- The total bioavailable fraction of Cd and Zn (2.6 – 5.1% and 0.32 – 0.61%, of the total Cd and Zn, respectively) was almost comparable for all the sensors whereas the bioavailability of Hg in soil-water suspensions was about 10 fold higher for Gram-negative sensor cells (30.5% of total Hg) compared to Gram-positive ones (3.2% of the total Hg).
- In the case of Zn, the water-extracted and total bioavailable fractions were equal indicating that no additional Zn could be mobilized by the sensor bacteria upon direct contact with soil matrix in suspension assay.
- The bioavailable fraction of Cd and Hg (only in the case of Gram-negative sensor strains) in soil-water suspensions exceeded the water-extracted bioavailable fraction about 14-fold indicating that upon direct contact, additional fraction of Cd and Hg was mobilized by those sensor bacteria
- Using Cd as a model we showed that in the used test conditions, 2-hour incubation (standard incubation time for the test with sensor bacteria) was enough for all the used bacterial strains to access all potentially available Cd in the soil-water suspensions.
Acknowledgments
References
- Bioavailability of Contaminants in Soils and Sediments: Processes, Tools, and Applications; The National Academies Press: Washington, D. C., 2003.
- Mountouris, A.; Voutsas, E.; Tassios, D. Bioconcentration of heavy metals in aquatic environments: the importance of bioavailability. Mar. Pollut. Bull. 2002, 44, 1136–1141. [Google Scholar]
- McKenzie, R.M. The adsorption of lead and other heavy metals on oxides of manganese and iron. Aust. J. Soil Res 1980, 18, 61–73. [Google Scholar]
- Martinez, C.E.; McBride, M.B.; Murray, B. Dissolved and labile concentrations of Cd, Cu, Pb, and Zn in aged ferrihydrite-organic matter systems. Environ. Sci. Technol. 1999, 33, 745–750. [Google Scholar]
- Rensing, C.; Maier, M. Issues underlying use of biosensors to measure metal bioavailability. Ecotoxicol. Environ. Saf. 2003, 56, 140–147. [Google Scholar]
- De Haan, F.A.M.; van Riemsdijk, W.H.; van der Zee, S.E. General concepts of soil quality. In Integrated Soil and Sediment Research: A Basis for Proper Protection.; Eijsackers, H.G.P., Hamers, T., Eds.; Kluwer Academic Publishers: Dordrecht, 1993; pp. 155–170. [Google Scholar]
- Semple, K.T.; Doick, K.J.; Jones, K.C.; Burauel, P.; Craven, A.; Harms, H. Defining bioavailability and bioaccessibility of contaminated soil and sediment is complicated. Environ. Sci. Technol. 2004, 38, 228A–231A. [Google Scholar]
- Vanhala, P.T.; Ahtiainen, J.H. Soil respiration, ATP content, and Photobacterium toxicity test as indicators of metal pollution in soil. Environ. Toxicol. Water Qual. 1994, 9, 115–121. [Google Scholar]
- Kahru, A.; Ivask, A.; Kasemets, K.; Põllumaa, L.; Kurvet, I.; Francois, M.; Dubourguier, H.C. Biotests and biosensors in ecotoxicological risk assessment of field soils polluted with zinc, lead and cadmium. Environ. Toxicol. Chem. 2005, 24, 2973–2982. [Google Scholar]
- Liao, V.H.C.; Chien, M.T.; Tseng, Y.Y.; Ou, K.L. Assessment of heavy metal bioavailability in contaminated sediments and soils using green fluorescent protein-based bacterial biosensors. Environ. Pollut. 2006, 142, 17–23. [Google Scholar]
- Ivask, A.; Virta, M.; Kahru, A. Construction and use of specific luminescent recombinant bacterial sensors for the assessment of bioavailable fraction of cadmium, zinc, mercury and chromium in the soil. Soil Biol. Biochem. 2002, 34, 1439–1447. [Google Scholar]
- Petänen, T.; Romantschuk, M. Toxicity and bioavailability to bacteria of article-associated arsenite and mercury. Chemosphere 2003b, 50, 409–413. [Google Scholar]
- Ivask, A.; Francois, M.; Kahru, A.; Dubourguier, H.C.; Virta, M.; Douay, F. Recombinant luminescent bacterial sensors for the measurement of bioavailability of cadmium and lead in soils polluted by metal smelters. Chemosphere 2004, 22, 147–156. [Google Scholar]
- Tipping, E.; Rieuwerts, J.; Pan, G.; Ashmore, M.R.; Lofts, S.; Hill, M.T.; Farago, M.E.; Thornton, I. The solid-solution partitioning of heavy metals (Cu, Zn, Cd, Pb) in upland soils of England and Wales. Environ. Pollut. 2003, 125, 213–225. [Google Scholar]
- Haferburg, G.; Kothe, E. Microbes and metals: interactions in the environment. J. Basic Microbiol. 2007, 47, 453–467. [Google Scholar]
- Gadd, G. M. Microbial influence on metal mobility and application for bioremediation. Geoderma 2004, 122, 109–119. [Google Scholar]
- Jiang, W.; Saxena, A.; Song., B.; Ward, B.B.; Beveridge, T.J. Myneni S.C.B. Elucidation of functional groups on Gram-positive and Gram-negative bacterial surfaces using infrared spectroscopy. Langmuir 2004, 20, 11433–11442. [Google Scholar]
- Beveridge, T.J.; Murray, R.G.E. Sites of metal deposition in the cell wall in Bacillus subtilis. J. Bacteriol. 1980, 141, 876–887. [Google Scholar]
- Beveridge, T.J. Ultrastructure, chemistry and function of the bacterial cell. Int. Rev. Cytol. 1981, 72, 229–317. [Google Scholar]
- Nies, D. Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol. Rev. 2003, 27, 313–339. [Google Scholar]
- Ure, A.M.; Davidson, C.M.; Thomas, R.P. Single and sequential extraction schemes for trace metal speciation in soil and sediment. In Quality Assurance for Environmental Analysis; Ph Quevauviller, E.A., Maier, B.G., Eds.; Elsevier: Belgium, 1995; Volume 20, pp. 505–523. [Google Scholar]
- Basta, N.; Gradwohl, R. Estimation of Cd, Pb, and Zn Bioavailability in Smelter-Contaminated Soils by a Sequential Extraction Procedure. J. Soil Contam. 2000, 9, 149–164. [Google Scholar]
- Harms, H.; Wells, M.; van der Meer, J.R. Whole-cell living biosensors - are they ready for environmental application? Appl. Microbiol. Biotechnol. 2006, 70, 272–280. [Google Scholar]
- Köhler, S.; Belkin, S.; Schmid, R.D. Reporter gene bioassays in environmental analysis. Fresenius. J. Anal. Chem. 2000, 366, 769–779. [Google Scholar]
- Daunert, S.; Barrett, G.; Feliciano, J.S.; Shetty, R.; Shrestha, S.; Smith-Spencer, W. Genetically engineered whole-cell sensing systems: coupling biological recognition with reporter genes. Chem. Rev. 2000, 100, 2705–2738. [Google Scholar]
- Ivask, A.; Green, T.; Polyak, B.; Mor, A.; Kahru, A.; Virta, M.; Marks, R. Fibre-optic bacterial biosensors and their application for the analysis of bioavailable Hg and As in soils and sediments from Aznalcollar mining area in Spain. Biosens. Bioelectron. 2007, 22, 1396–1402. [Google Scholar]
- Corbisier, P.; Thiry, E.; Diels, L. Bacterial biosensors for the toxicity assessment of solid wastes. Environ. Toxicol. Water Qual. 1996, 11, 171–177. [Google Scholar]
- Peltola, P.; Ivask, A.; Åström, M.; Virta, M. Lead and Cu in contaminated urban soils: Extraction with chemical reagents and bioluminescent bacteria and yeast. Sci. Total Environ. 2005, 350, 194–203. [Google Scholar]
- Brandt, K.K.; Holm, P.E.; Nybroe, O. Bioavailability and toxicity of soil particle-associated copper as determined by two bioluminescent Pseudomonas fluorescens biosensor strains. Environ. Toxicol. Chem. 2006, 25, 1738–1741. [Google Scholar]
- Rasmussen, L.D.; Sorensen, S.J.; Turner, R.R.; Barkay, T. Application of a mer-lux biosensor for estimating bioavailable mercury in soil. Soil Biol. Biochem. 2000, 32, 639–46. [Google Scholar]
- Turpeinen, R.; Salminen, J.; Kairesalo, T. Mobility and bioavailability of lead in contaminated boreal forest soil. Environ. Sci. Technol. 2000, 34, 5152–5156. [Google Scholar]
- Fritze, H.; Perikomäki, J.; Petänen, T.; Pennanen, T.; Romantschuk, M.; Karp, M.; Yrjälä, K. A microcosmos study on the effects of Cd-containing wood ash on the coniferous humus fungal community and the Cd bioavailability. J. Soils Sedim. 2001, 1, 146–150. [Google Scholar]
- Tibazarwa, C.; Corbisier, P.; Mench, M.; Bossus, A.; Solda, P.V.; Mergeay, M.; Wyns, L.; van der Lelie, D. A microbial biosensor to predict bioavailable nickel and its transfer to plants. Environ. Pollut. 2001, 113, 19–26. [Google Scholar]
- Petänen, T.; Lyytikainen, M.; Lappalainen, J.; Romantschuk, M.; Kukkonen, J.V.K. Assessing sediment toxicity and arsenite concentration with bacterial and traditional methods. Environ. Pollut. 2003, 122, 407–415. [Google Scholar]
- Heinlaan, M.; Ivask, A.; Blinova, I.; Dubourguier, H.C.; Kahru, A. Toxicity of nanosized and bulk ZnO, CuO and TiO2 to bacteria Vibrio fischeri and crustaceans Daphnia magna and Thamnocephalus platyurus. Chemosphere 2008, 71, 1308–1316. [Google Scholar]
- Kahru, A.; Dubourguier, H.C.; Blinova, I.; Ivask, A.; Kasemets, K. Biotests and biosensors for ecotoxicology of metal oxide nanoparticles: a minireview. Sensors 2008, 8, 5153–5170. [Google Scholar]
- Ivask, A.; Hakkila, K.; Virta, M. Detection of organomercurials with sensor bacteria. Anal. Chem. 2001, 73, 5168–5171. [Google Scholar]
- Ivask, A.; Rõlova, T.; Bondareko, O.; Virta, M.; Kahru, A. A suite of recombinant luminescent bacterial strains for the quantification of bioavailable heavy metals and toxicity testing. BMC Biotech. Submitted.
- Hakkila, K.; Maksimow, M.; Karp, M.; Virta, M. Reporter genes lucFF, luxCDABE, gfp and dsred have different characteristics in whole-cell bacterial sensors. Anal Biochem 2002, 301, 235–242. [Google Scholar]
- Madigan, M.; Martinko, J. (Eds.) Brock Biology of Microorganisms, 11th ed.; Prentice Hall: St. Paul, 2005.
- Nies, D. Bacterial Transition Metal Homeostasis. In Molecular Microbiology of Heavy Metals; Nies, D., Silver, S., Eds.; Springer: Berlin/Heidelberg, 2007; pp. 118–137. [Google Scholar]
- Dittrich, C.R.; Bennett, G.N.; San, K.Y. Characterization of the acetate-producing pathways in. Escherichia coli. Biotechnol. Progr. 2005, 21, 1062–1067. [Google Scholar]
- Ochoa-Loza, F.J.; Artiola, J.F.; Maier, R.M. Stability constants for the complexation of various metals with a rhamnolipid biosurfactant. J. Environ. Qual. 2001, 30, 479–485. [Google Scholar]
- Juwarkar, A.A.; Nair, A.; Dubey, K.V; Singh, S.K.; Devotta, S. Biosurfactant technology for remediation of cadmium and lead contaminated soils. Chemosphere 2007, 68, 1996–2002. [Google Scholar]
- Moriarty, D.J.W.; Hayward, A.C. Ultrastructure of bacteria and the proportion of Gram-negative bacteria in marine sediments. Microb. Ecol. 1982, 8, 1–14. [Google Scholar]
- Baldi, F. Microbial transformation of mercury species and their importance in the biogeochemical cycle of mercury. In Mercury and its effects on Environment and Biology; Sigel, H., Sigel, A., Eds.; Marcel Dekker, Inc.: New York, 1996; pp. 213–257. [Google Scholar]
- Harms, H.; Bosma, T.N.P. Mass transfer limitation of microbial growth and pollutant degradation. J. Ind. Microbiol. Biotechnol. 1997, 18, 97–110. [Google Scholar]
- Sarand, I.; Haario, H.; Jorgenson, K.S.; Romantshuk, M. Effect of inoculation of a TOL plasmid containing mycorrhizosphere bacterium on development of Scots pine seedlings, their mycorrhizosphere and the microbial flora in m-toluate-amended soil. FEMS Microbiol. Ecol. 2000, 31, 127–141. [Google Scholar]
- Frackman, S.; Anhalt, M.; Nealson, K.H. Cloning, organization, and expression of the bioluminescence genes of Xenorhabdus luminescens. J. Bacteriol. 1990, 172, 5767–5773. [Google Scholar]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular cloning. In A laboratory manual.; Cold Spring Harbour Laboratory Press: New York, 1989. [Google Scholar]
- Hakkila, K.; Green, T.; Leskinen, P.; Ivask, A.; Marks, R.; Virta, M. Detection of bioavailable heavy metals in EILATox-Oregon samples using whole-cell luminescent bacterial sensors in suspension or immobilized onto Fibre-optic Tips. J. Appl. Toxicol. 2004, 24, 333–342. [Google Scholar]





| Host bacterium and sensor strain | Genetic metal-response element a | Location of metal-response elements | Inducing metals b | Limit of determination (LOD), μg·L-1±SD | |||
|---|---|---|---|---|---|---|---|
| Cd2+ | Zn2+ | Hg2+ | |||||
| Gram-negative | |||||||
| Pseudomonas fluorescensOS8 | |||||||
| OS8::KncadRPcadAlux c | CadR/PcadA | chromosome | Cd, Zn, Hg,Pb | 7 ± 2.5 | 400 ± 110 | 4 ± 1.3 | |
| OS8(pDNcadRPcadAlux)d | CadR/PcadA | plasmid | Cd, Zn, Hg,Pb | 8 ± 1.4 | 500 ± 40 | 15 ± 1.7 | |
| OS8::KnzntRPzntAluxc | ZntR/PzntA | chromosome | Cd, Zn, Hg,Pb | 20 ± 4.7 | 5000 ± 580 | 60 ± 19 | |
| OS8::KnmerRBSBPmerluxc | MerB/MerR/Pmer | chromosome | Hg, MeHg, Cd | 4500 ± 1230 | not induced | 0.8 ± 0.2 | |
| OS8(pDNmerRBSBPmerlux)d | MerB/MerR/Pmer | plasmid | Hg, MeHg, Cd | 650 ± 220 | not induced | 0.2 ± 0.05 | |
| Escherichia coli MC1061 | |||||||
| MC1061(pSLzntR/pDNPzntAlux)d | ZntR/PzntA | plasmid | Cd, Zn, Pb | 2 ± 0.5 | 700 ± 170 | 20 ± 5 | |
| MC1061 (pmerRBSBPmerlux)d | MerB/MerR/Pmer | plasmid | Hg, MeHg, Cd | 40 ± 13 | not induced | 0.03 ± 0.009 | |
| MC1061(pmerGFP)f | MerR/Pmer | plasmid | Hge | 0.6e | |||
| Gram-positive | |||||||
| Bacillus subtilisBR151 | |||||||
| BR151(pcadCPcadAlux)d | CadC/PcadA | plasmid | Cd, Zn, Hg,Pb | 2 ±0.3 | 1000 ± 150 | 10 ± 1.5 | |
| Staphylococcus aureusRN4220 | |||||||
| RN4220(pcadCPcadAlux)d | CadC/PcadA | plasmid | Cd, Zn, Hg,Pb | 7 ± 2 | 1500 ± 210 | 2 ± 0.7 | |
| Metal | Host bacterium (species) | Strain | Metal-response elements (location)a | Viable cells in test | Total bioavailable b,%of total | Water-extracted bioavailablec,%of total |
|---|---|---|---|---|---|---|
| Cdd | Gram-negative | |||||
| Pseudomonas fluorescens | OS8::KncadR PcadAlux | CadR/PcadA (C) | 3×107 | 3.5 ± 1.8 | 0.46 ± 0.19 | |
| OS8(pDNcadR PcadAlux) | CadR/PcadA (P) | 4×106 | 4.4 ± 2.5 | ND | ||
| OS8::KnzntR PzntAlux | ZntR/PzntA (C) | 6×106 | 4.8 ± 0.7 | 0.23 ± 0.0011 | ||
| OS8::KnmerRBSB Pmerlux | MerR/Pmer (C) | 2×107 | 2.6 ± 0.4 | 0.41 ± 0.039 | ||
| OS8(pDNmerRBSB Pmerlux) | MerR/Pmer (P) | 5×106 | 3.7 ± 1.5 | ND | ||
| Escherichia coli | MC1061(pSLzntR/ pDNPzntAlux) | ZntR/PzntA (P) | 1×107 | 5.1 + 0.51 | 0.24 ± 0.18 | |
| MC1061(pmerRBSB Pmerlux) | MerR/Pmer (P) | 4×107 | 3.7 ± 1.5 | 0.44 ± 0.18 | ||
| AVERAGE for Gram-negative bacteria | 4.2±0.71 | 0.36±0.11 | ||||
| Gram-positive | ||||||
| Bacillus subtilis | BR151(pcadC PcadAlux) | CadC/PcadA (P) | 3×106 | 3.2 ± 1.1 | 0.19 ± 0.08 | |
| Staphylococcus aureus | RN4220(pcadC PcadAlux) | CadC/PcadA (P) | 8×106 | 2.6 ± 0.3 | 0.38 ± 0.054 | |
| AVERAGE for Gram-positive bacteria | 2.9±0.36 | 0.28±0.13 | ||||
| Hg e | Gram-negative | |||||
| Pseudomonas fluorescens | OS8::KncadR PcadAlux | CadR/PcadA (C) | 3×107 | 27.0 ± 7.7 | 2.4 ± 0.73 | |
| OS8(pDNcadR PcadAlux) | CadR/PcadA (P) | 4×106 | 28.1 ± 14.0 | ND | ||
| OS8::KnzntR PzntAlux | ZntR/PzntA (C) | 6×106 | 26.7 ± 1.1 | 2.6 ± 0.16 | ||
| OS8::KnmerRBSB Pmerlux | MerR/Pmer (C) | 2×107 | 31.9 ± 12.2 | 2.6 ± 0.58 | ||
| OS8(pDNmerRBSB Pmerlux) | MerR/Pmer (P) | 5×106 | 18.7 ± 7.3 | ND | ||
| Escherichia coli | MC1061(pmerRBSB Pmerlux) | MerR/Pmer (P) | 1×107 | 27.9 ± 5.3 | 1.9 | |
| MC1061(pSLzntR/ pDNPzntAlux) | ZntR/PzntA (P) | 4×107 | 38.9 ± 5.6 | 1.67 ± 0.40 | ||
| AVERAGE for Gram-negative bacteria | 30.5±5.17 | 2.2±0.43 | ||||
| Hg e | Gram-positive | |||||
| Bacillus subtilis | BR151(pcadC PcadAlux) | CadC/PcadA (P) | 3×106 | 3.8 ± 2.8 | 4.9 ± 1.3 | |
| Staphylococcus aureus | RN4220(pcadCPcadAlux) | CadC/PcadA (P) | 8×106 | 2.6 ± 1.2 | 2.8 ± 1.3 | |
| AVERAGE for Gram-positive bacteria | 3.22±0.81 | 3.92±1.51 | ||||
| Znf | Gram-negative | |||||
| Pseudomonas fluorescens | OS8::KncadR PcadAlux | CadR/PcadA (C) | 3×107 | 0.35 ± 0.17 | 0.34 ± 0.18 | |
| OS8(pDNcadR PcadAlux) | CadR/PcadA (P) | 4×106 | 0.32 ± 0.045 | ND | ||
| OS8::KnzntR PzntAlux | ZntR/PzntA (C) | 6×106 | 0.42 ± 0.19 | 0.37 + 0.11 | ||
| Escherichia coli | MC1061(pSLzntR/ pDNPzntAlux) | ZntR/PzntA (P) | 4×107 | 0.61 ± 0.35 | 0.27 ± 0.037 | |
| AVERAGE for Gram-negative bacteria | 0.36±0.05 | 0.37±0.04 | ||||
| Gram-positive | ||||||
| Bacillus subtilis | BR151(pcadC PcadAlux) | CadC/PcadA (P) | 3×106 | 0.44 ± 0.20 | 0.24 ± 0.11 | |
| Staphylococcus aureus | RN4220(pcadCPcadAlux) | CadC/PcadA (P) | 8×106 | 0.33 ± 0.18 | 0.25 ± 0.0018 | |
| AVERAGE for Gram-positive bacteria | 0.39±0.07 | 0.25±0.004 | ||||
| Bacterium | Time of incubation, min | |||
|---|---|---|---|---|
| 0 | 30 | 60 | 120 | |
| Pseudomonas fluorescens 0S8::KncadRPcadAlux | 1.7 | 2.21 | 2.19 | 2.21 |
| Escherichia coli MCI061 (pSLzntR/pDNPzntAlux) | 1.7 | 3.18 | 3.20 | 3.19 |
| Staphylococcus aureus RN4220(pcadCPcadAlux) | 1.7 | 2.60 | 2.50 | 2.56 |
| Bacillus subtilis BR151 (pcadCPcadAlux) | 1.7 | 2.70 | 2.85 | 3.41 |
| Soil | Cd | Zn | Hg |
|---|---|---|---|
| Total, mg·kg1dwt of soil | |||
| Non-spiked soila | 0.45 | 219 | 0.14 |
| Spiked soil samples | Added metalb, mg·_kg-1dwt of soil | ||
| 1 | 1.5 | 0 | 0 |
| 2 | 15 | 0 | 0 |
| 3 | 150 | 0 | 0 |
| 4 | 1500 | 0 | 0 |
| 5 | 15000 | 0 | 0 |
| 6 | 0 | 900 | 0 |
| 7 | 0 | 9000 | 0 |
| 8 | 0 | 90000 | 0 |
| 9 | 0 | 0 | 0.28 |
| 10 | 0 | 0 | 2.8 |
| 11 | 0 | 0 | 17 |
| 12 | 0 | 0 | 28 |
| 13 | 0 | 0 | 280 |
| Permitted limit values for soilc | |||
| 1-3 | 150-300 | 1-1.5 | |
© 2008 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Bondarenko, O.; Rõlova, T.; Kahru, A.; Ivask, A. Bioavailability of Cd, Zn and Hg in Soil to Nine Recombinant Luminescent Metal Sensor Bacteria. Sensors 2008, 8, 6899-6923. https://doi.org/10.3390/s8116899
Bondarenko O, Rõlova T, Kahru A, Ivask A. Bioavailability of Cd, Zn and Hg in Soil to Nine Recombinant Luminescent Metal Sensor Bacteria. Sensors. 2008; 8(11):6899-6923. https://doi.org/10.3390/s8116899
Chicago/Turabian StyleBondarenko, Olesja, Taisia Rõlova, Anne Kahru, and Angela Ivask. 2008. "Bioavailability of Cd, Zn and Hg in Soil to Nine Recombinant Luminescent Metal Sensor Bacteria" Sensors 8, no. 11: 6899-6923. https://doi.org/10.3390/s8116899