pMPES: A Modular Peptide Expression System for the Delivery of Antimicrobial Peptides to the Site of Gastrointestinal Infections Using Probiotics
Abstract
:1. Introduction
2. Results
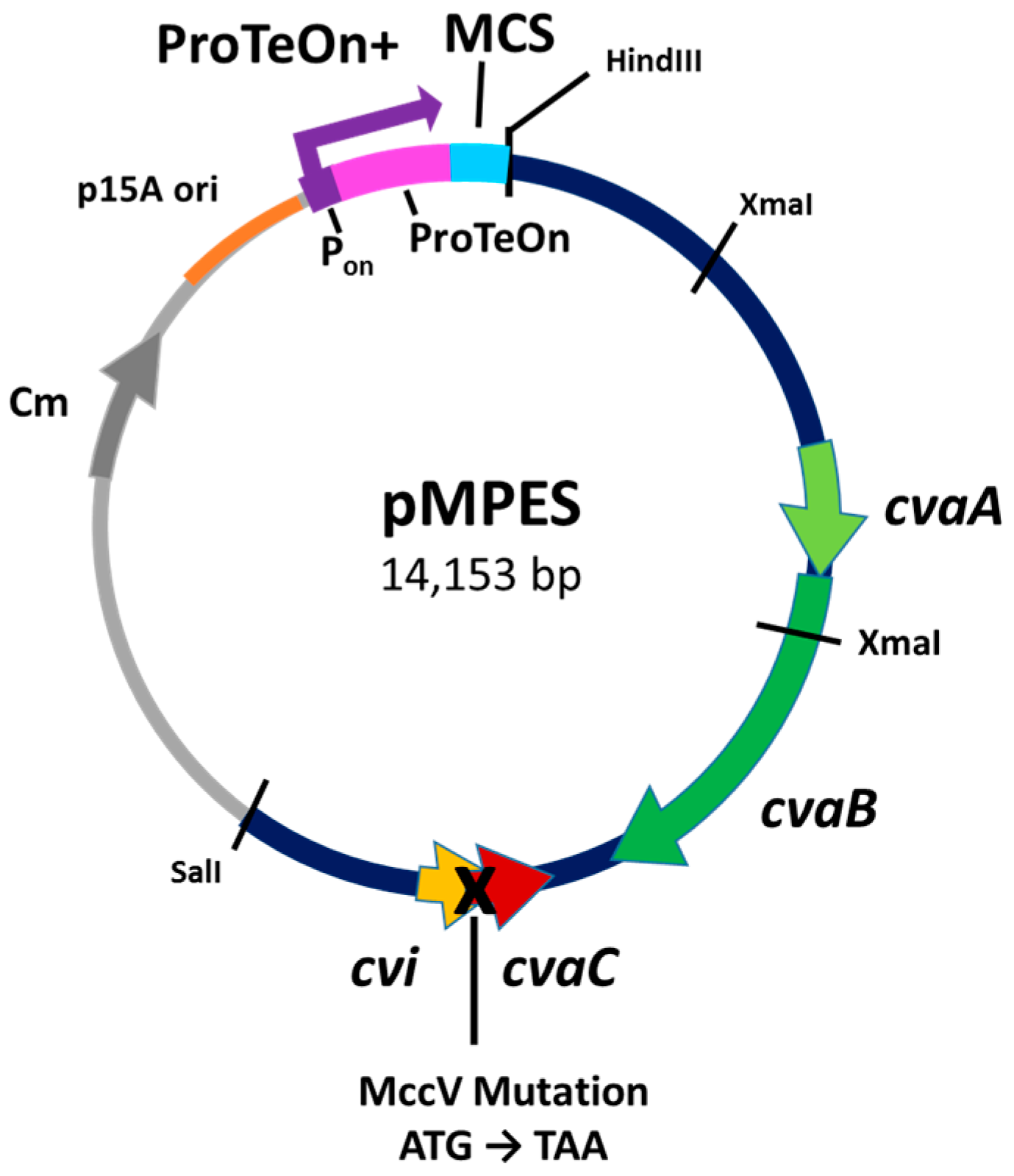
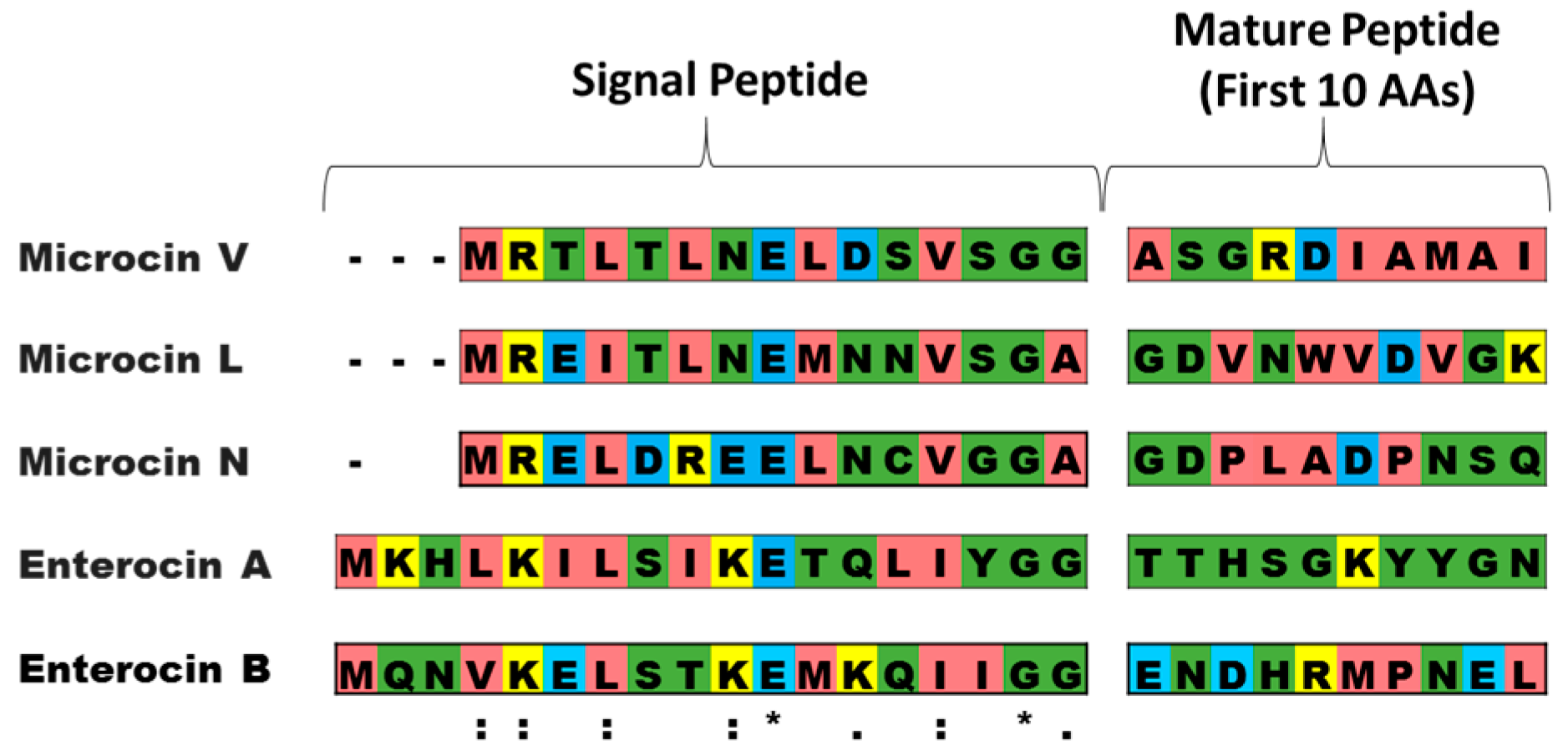
2.1. Development of the Vector
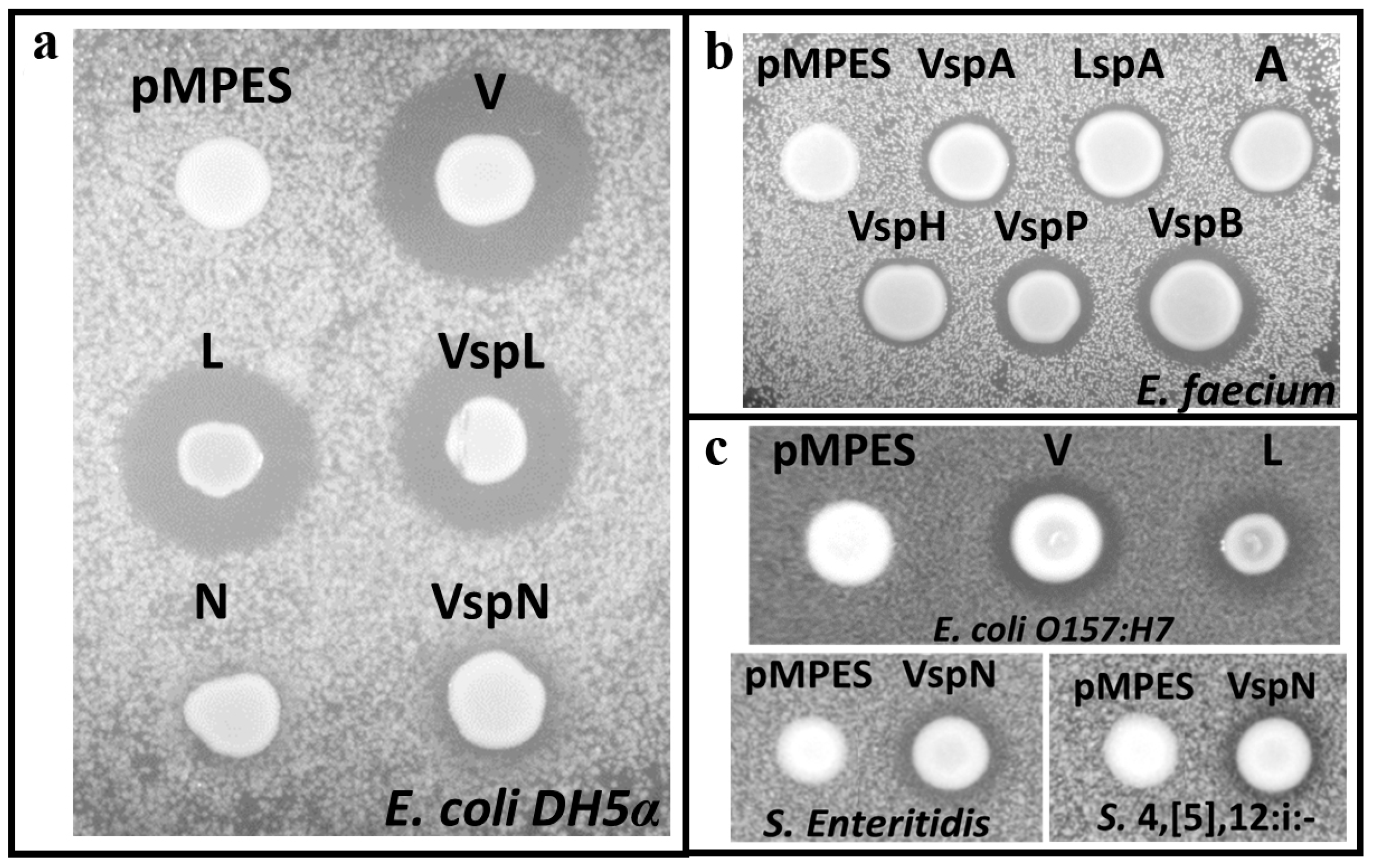
2.2. Activity Tests
2.3. Simultaneous Expression of Multiple AMPs
2.4. Supernatant Activity Assays
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Plasmids
Bacteria Growth Conditions
4.2. Construction of Plasmids
4.3. Activity Assays
4.3.1. Agar Diffusion Assays
4.3.2. Supernatant Activity Assays
4.4. Peptide Isolation Attempts
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Geldart, K.; Borrero, J.; Kaznessis, Y.N. A chloride-inducible expression vector for delivery of antimicrobial peptides against antibiotic-resistant enterococcus faecium. Appl. Environ. Microbiol. 2015, 81, 3889–3897. [Google Scholar] [CrossRef] [PubMed]
- Destoumieux-Garzón, D.; Duquesne, S.; Peduzzi, J.; Goulard, C.; Desmadril, M.; Letellier, L.; Rebuffat, S.; Boulanger, P. The iron-siderophore transporter FhuA is the receptor for the antimicrobial peptide microcin J25: Role of the microcin Val11-Pro16 beta-hairpin region in the recognition mechanism. Biochem. J. 2005, 389, 869–876. [Google Scholar] [CrossRef] [PubMed]
- Cintas, L.M.; Casaus, P.; Håvarstein, L.S.; Hernández, P.E.; Nes, I.F. Biochemical and genetic characterization of enterocin P, a novel sec-dependent bacteriocin from Enterococcus faecium P13 with a broad antimicrobial spectrum. Appl. Environ. Microbiol. 1997, 63, 4321–4330. [Google Scholar] [PubMed]
- Khandelia, H.; Langham, A.A.; Kaznessis, Y.N. Driving engineering of novel antimicrobial peptides from simulations of peptide–micelle interactions. Biochim. Biophys. Acta Biomembr. 2006, 1758, 1224–1234. [Google Scholar] [CrossRef] [PubMed]
- Oelschlaeger, T.A. Mechanisms of probiotic actions—A review. Int. J. Med. Microbiol. 2010, 300, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Cotter, P.D.; Ross, R.P.; Hill, C. Bacteriocins—A viable alternative to antibiotics? Nat. Rev. Microbiol. 2013, 11, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Gillor, O.; Etzion, A.; Riley, M.A. The dual role of bacteriocins as anti- and probiotics. Appl. Microbiol. Biotechnol. 2008, 81, 591–606. [Google Scholar] [CrossRef] [PubMed]
- Borrero, J.; Jiménez, J.J.; Gútiez, L.; Herranz, C.; Cintas, L.M.; Hernández, P.E. Protein expression vector and secretion signal peptide optimization to drive the production, secretion, and functional expression of the bacteriocin enterocin A in lactic acid bacteria. J. Biotechnol. 2011, 156, 76–86. [Google Scholar] [CrossRef] [PubMed]
- Cano-Garrido, O.; Seras-Franzoso, J.; Garcia-Fruitós, E. Lactic acid bacteria: reviewing the potential of a promising delivery live vector for biomedical purposes. Microb. Cell Fact. 2015, 14, 137. [Google Scholar] [CrossRef] [PubMed]
- Braat, H.; Rottiers, P.; Hommes, D.W.; Huyghebaert, N.; Remaut, E.; Remon, J.-P.; van Deventer, S.J.H.; Neirynck, S.; Peppelenbosch, M.P.; Steidler, L. A phase I trial with transgenic bacteria expressing interleukin-10 in Crohn’s disease. Clin. Gastroenterol. Hepatol. 2006, 4, 754–759. [Google Scholar] [CrossRef] [PubMed]
- Schultz, M.D. Clinical use of E. coli Nissle 1917 in inflammatory bowel disease. Inflamm. Bowel Dis. 2008, 14, 1012–1018. [Google Scholar] [CrossRef] [PubMed]
- Bryan, E.M.; Bae, T.; Kleerebezem, M.; Dunny, G.M. Improved Vectors for Nisin-Controlled Expression in Gram-Positive Bacteria. Plasmid 2000, 44, 183–190. [Google Scholar] [CrossRef] [PubMed]
- De Vos, W.M. Gene expression systems for lactic acid bacteria. Curr. Opin. Microbiol. 1999, 2, 289–295. [Google Scholar] [CrossRef]
- Borrero, J.; Chen, Y.; Dunny, G.M.; Kaznessis, Y.N. Modified lactic acid bacteria detect and inhibit multiresistant enterococci. ACS Synth. Biol. 2015, 4, 299–306. [Google Scholar] [CrossRef] [PubMed]
- Volzing, K.; Borrero, J.; Sadowsky, M.J.; Kaznessis, Y.N. Antimicrobial peptides targeting Gram-negative pathogens, produced and delivered by lactic acid bacteria. ACS Synth. Biol. 2013, 2, 643–650. [Google Scholar] [CrossRef] [PubMed]
- Forkus, B.; Vlysidis, M.; Ritter, S.; Geldart, K.; Kaznessis, Y.N. Antimicrobial probiotics reduce salmonella enterica in Turkey gastrointestinal tracts. 2016; submitted. [Google Scholar]
- O’Keeffe, T.; Hill, C.; Ross, R.P. Characterization and heterologous expression of the genes encoding enterocin a production, immunity, and regulation in Enterococcus faecium DPC1146. Appl. Environ. Microbiol. 1999, 65, 1506–1515. [Google Scholar] [PubMed]
- Gilson, L.; Mahanty, H.K.; Kolter, R. Four plasmid genes are required for colicin V synthesis, export, and immunity. J. Bacteriol. 1987, 169, 2466–2470. [Google Scholar] [PubMed]
- Pons, A.-M.; Delalande, F.; Duarte, M.; Benoit, S.; Lanneluc, I.; Sablé, S.; Van Dorsselaer, A.; Cottenceau, G. Genetic analysis and complete primary structure of microcin L. Antimicrob. Agents Chemother. 2004, 48, 505–513. [Google Scholar] [CrossRef] [PubMed]
- Azpiroz, M.F.; Rodríguez, E.; Laviña, M. The structure, function, and origin of the microcin H47 ATP-binding cassette exporter indicate its relatedness to that of colicin V. Antimicrob. Agents Chemother. 2001, 45, 969–972. [Google Scholar] [CrossRef] [PubMed]
- Aucher, W.; Lacombe, C.; Hequet, A.; Frere, J.; Berjeaud, J.-M. Influence of amino acid substitutions in the leader peptide on maturation and secretion of mesentericin Y105 by leuconostoc mesenteroides. J. Bacteriol. 2005, 187, 2218–2223. [Google Scholar] [CrossRef] [PubMed]
- Van Belkum, M.J.; Worobo, R.W.; Stiles, M.E. Double-glycine-type leader peptides direct secretion of bacteriocins by ABC transporters: colicin V secretion in Lactococcus lactis. Mol. Microbiol. 1997, 23, 1293–1301. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, J.; Borrero, J.; Gómez-Sala, B.; Basanta, A.; Herranz, C.; Cintas, L.M.; Hernández, P.E. Cloning and heterologous production of Hiracin JM79, a sec-dependent bacteriocin produced by Enterococcus hirae DCH5, in lactic acid bacteria and Pichia pastoris. Appl. Environ. Microbiol. 2008, 74, 2471–2479. [Google Scholar] [CrossRef] [PubMed]
- Jiménez, J.J.; Diep, D.B.; Borrero, J.; Gútiez, L.; Arbulu, S.; Nes, I.F.; Herranz, C.; Cintas, L.M.; Hernández, P.E. Cloning strategies for heterologous expression of the bacteriocin enterocin A by Lactobacillus sakei Lb790, Lb. plantarum NC8 and Lb. casei CECT475. Microb. Cell Fact. 2015, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klocke, M.; Mundt, K.; Idler, F.; Jung, S.; Backhausen, J.E. Heterologous expression of enterocin A, a bacteriocin from Enterococcus faecium, fused to a cellulose-binding domain in Escherichia coli results in a functional protein with inhibitory activity against Listeria. Appl. Microbiol. Biotechnol. 2005, 67, 532–538. [Google Scholar] [CrossRef] [PubMed]
- McCormick, J.K.; Worobo, R.W.; Stiles, M.E. Expression of the antimicrobial peptide carnobacteriocin B2 by a signal peptide-dependent general secretory pathway. Appl. Environ. Microbiol. 1996, 62, 4095–4099. [Google Scholar] [PubMed]
- Gilson, L.; Mahanty, H.K.; Kolter, R. Genetic analysis of an MDR-like export system: The secretion of colicin V. EMBO J. 1990, 9, 3875–3884. [Google Scholar] [PubMed]
- Skvirsky, R.C.; Shen, X.; Reginald, S. Is the CvaA* protein, encoded within the colicin V export gene cvaA, required for colicin V transport? FEMS Microbiol. Lett. 1996, 138, 201–206. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.H.; Fath, M.J.; Mahanty, H.K.; Tai, P.C.; Kolter, R. Genetic analysis of the colicin V secretion pathway. Genetics 1995, 141, 25–32. [Google Scholar] [PubMed]
- Hwang, J.; Zhong, X.; Tai, P.C. Interactions of dedicated export membrane proteins of the colicin V secretion system: CvaA, a member of the membrane fusion protein family, interacts with CvaB and ToIC. J. Bacteriol. 1997, 179, 6264–6270. [Google Scholar] [PubMed]
- Volzing, K.; Biliouris, K.; Kaznessis, Y.N. proTeOn and proTeOff, new protein devices that inducibly activate bacterial gene expression. ACS Chem. Biol. 2011, 6, 1107–1116. [Google Scholar] [CrossRef] [PubMed]
- Corsini, G.; Karahanian, E.; Tello, M.; Fernandez, K.; Rivero, D.; Saavedra, J.M.; Ferrer, A. Purification and characterization of the antimicrobial peptide microcin N. FEMS Microbiol. Lett. 2010, 312, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Casaus, P.; Nilsen, T.; Cintas, L.M.; Nes, I.F.; Hernández, P.E.; Holo, H. Enterocin B, a new bacteriocin from Enterococcus faecium T136 which can act synergistically with enterocin A. Microbiology 1997, 143, 2287–2294. [Google Scholar] [CrossRef] [PubMed]
- Pons, A.-M.; Lanneluc, I.; Cottenceau, G.; Sable, S. New developments in non-post translationally modified microcins. Biochimie 2002, 84, 531–537. [Google Scholar] [CrossRef]
- Kaur, K.; Tarassova, O.; Dangeti, R.V.; Azmi, S.; Wishart, D.; McMullen, L.; Stiles, M. Characterization of a highly potent antimicrobial peptide microcin N from uropathogenic Escherichia coli. FEMS Microbiol. Lett. 2016, 363. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, J.; Diep, D.B.; Herranz, C.; Nes, I.F.; Cintas, L.M.; Hernández, P.E. Amino acid and nucleotide sequence, adjacent genes, and heterologous expression of hiracin JM79, a sec-dependent bacteriocin produced by Enterococcus hirae DCH5, isolated from Mallard ducks (Anas platyrhynchos). FEMS Microbiol. Lett. 2007, 270, 227–236. [Google Scholar] [CrossRef] [PubMed]
- Franz, C.M.A.P.; Worobo, R.W.; Quadri, L.E.N.; Schillinger, U.; Holzapfel, W.H.; Vederas, J.C.; Stiles, M.E. Atypical Genetic Locus Associated with Constitutive Production of Enterocin B by Enterococcus faecium BFE 900. Appl. Environ. Microbiol. 1999, 65, 2170–2178. [Google Scholar] [PubMed]
- Rice, P.; Longden, I.; Bleasby, A. EMBOSS: The european molecular biology open software suite. Trends Genet. 2000, 16, 276–277. [Google Scholar] [CrossRef]
- Choo, K.H.; Ranganathan, S. Flanking signal and mature peptide residues influence signal peptide cleavage. BMC Bioinform. 2008, 9 (Suppl. 12), S15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]
- Soyer, Y.; Switt, M.A.; Davis, M.A.; Maurer, J.; McDonough, P.L.; Schoonmaker-Bopp, D.J.; Dumas, N.B.; Root, T.; Warnick, L.D.; Grohn, Y.T.; et al. Salmonella enterica Serotype 4,5,12:i:-, an Emerging Salmonella Serotype That Represents Multiple Distinct Clones. J. Clin. Microbiol. 2009, 47, 3546–3556. [Google Scholar] [CrossRef] [PubMed]
- Yeni, F.; Yavaş, S.; Alpas, H.; Soyer, Y. Most Common Foodborne Pathogens and Mycotoxins on Fresh Produce: A Review of Recent Outbreaks. Crit. Rev. Food Sci. Nutr. 2016, 56, 1532–1544. [Google Scholar] [CrossRef] [PubMed]
- Patzer, S.I.; Baquero, M.R.; Bravo, D.; Moreno, F.; Hantke, K. The colicin G, H and X determinants encode microcins M and H47, which might utilize the catecholate siderophore receptors FepA, Cir, Fiu and IroN. Microbiology 2003, 149, 2557–2570. [Google Scholar] [CrossRef] [PubMed]
- Salis, H.M. The ribosome binding site calculator. Methods Enzymol. 2011, 498, 19–42. [Google Scholar] [PubMed]
- Håvarstein, L.S.; Holo, H.; Nes, I.F. The leader peptide of colicin V shares consensus sequences with leader peptides that are common among peptide bacteriocins produced by Gram-positive bacteria. Microbiology 1994, 140, 2383–2389. [Google Scholar] [CrossRef] [PubMed]
- Morin, N.; Lanneluc, I.; Connil, N.; Cottenceau, M.; Pons, A.M.; Sablé, S. Mechanism of bactericidal activity of microcin L in Escherichia coli and Salmonella enterica. Antimicrob. Agents Chemother. 2011, 55, 997–1007. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.-B.; Malaphan, W.; Zendo, T.; Nakayama, J.; Sonomoto, K. Enterocin X, a novel two-peptide bacteriocin from Enterococcus faecium KU-B5, has an antibacterial spectrum entirely different from those of its component peptides. Appl. Environ. Microbiol. 2010, 76, 4542–4545. [Google Scholar] [CrossRef] [PubMed]
- Mathiesen, G.; Sveen, A.; Piard, J.-C.; Axelsson, L.; Eijsink, V.G.H. Heterologous protein secretion by Lactobacillus plantarum using homologous signal peptides. J. Appl. Microbiol. 2008, 105, 215–226. [Google Scholar] [CrossRef] [PubMed]
- Herranz, C.; Driessen, A.J.M. Sec-mediated secretion of bacteriocin enterocin P by Lactococcus lactis. Appl. Environ. Microbiol. 2005, 71, 1959–1963. [Google Scholar] [CrossRef] [PubMed]
- Ng, D.T.W.; Sarkar, C.A. Engineering signal peptides for enhanced protein secretion from Lactococcus lactis. Appl. Environ. Microbiol. 2013, 79, 347–356. [Google Scholar] [CrossRef] [PubMed]
- Azpiroz, M.F.; Laviña, M. Modular structure of microcin H47 and colicin V. Antimicrob. Agents Chemother. 2007, 51, 2412–2419. [Google Scholar] [CrossRef] [PubMed]
- Oswald, N. DIY Electrocompetent E. coli. Available online: http://bitesizebio.com/969/diy-electrocompetent-e-coli/ (accessed on 5 July 2016).
- E. coli Pulser TM Transformation Apparatus: Operating Instructions and Applications Guide. Available online: http://www.bio-rad.com/webroot/web/pdf/lsr/literature/M1652101C.pdf (accessed on 15 September 2016).


| AMP | Signal Peptide | Mature Peptide | Transporter (CvaB) | Accessory (CvaA) |
|---|---|---|---|---|
| MccL | 60%/86.7% | 44.1%/51.0% | 95.4%/97.3% | 97.6%/99.3% |
| McnN | 41.2%/47.1% | 20.8%/26.4% | 71.5%/85.0% | 69.4%/84.5% |
| EntA | 22.2%/50% | 10.3%/14.0% | 24.8%/43.4% | 18.5%/37.0% |
| EntB | 22.2%/50% | 14.9%/18.8% | NA | NA |
| HirJM79 | 10.0%/16.7% | 6.0%/6.9% | NA | NA |
| EntP | 6.5%/19.4% | 5.5%/11.0% | NA | NA |
| Construct (pMPES:) | Signal Peptide | Mature Peptide | Immunity Gene Included |
|---|---|---|---|
| V | MccV | MccV | Yes |
| L | MccL | MccL | Yes |
| VspL | MccV | MccL | Yes |
| N | McnN | McnN | Yes |
| VspN | MccV | McnN | Yes |
| VspA | MccV | EntA | Yes |
| LspA | MccL | EntA | Yes |
| A | EntA | EntA | Yes |
| VspH | MccV | HirJM79 | No |
| VspP | MccV | EntP | No |
| VspB | MccV | EntB | No |
| Indicator | Construct (pMPES:) | Bacteriocin Units (BU) 1 | Previously Reported MIC |
|---|---|---|---|
| E. coli DH5α | V | 120.9 ± 86.2 | 0.1 nM (E. coli MH1) [45] |
| L | >170.7 | 160 nM (E. coli ML-35p) [46] | |
| VL | 142.2 ± 49.3 | 160 nM (E. coli ML-35p) [46] | |
| N | 1.3 ± 0 | 150 nM (S. Enteriditis) [35] | |
| VspN | 1.3 ± 0 | 150 nM (S. Enteriditis) [35] | |
| E. faecium 8E9 | VspA | 7.2 ± 3.4 | 129 nM (E. faecium TUA 1344L) [47] |
| LspA | 7.2 ± 3.4 | 129 nM (E. faecium TUA 1344L) [47] | |
| A | 1.3 ± 0 | 129 nM (E. faecium TUA 1344L) [47] | |
| VspH | 21.3 ± 0 | ~0.2 nM (E. faecium T136) [23] | |
| VspP | 2.7 ± 0 | ~0.4 nM (E. faecium T136) [3] | |
| VspB | 2.7 ± 0 | 43.4 nM (E. faecium TUA 1344L ) [47] |
| Strain | Description | Source |
| E. coli MC1061 F' | Plasmid-free, recA+, non-amber suppressor strain | Lucigen |
| E. coli DH5α PRO | Derivative of E. coli DH5α; PN25/tetR, Placiq/laci, cloning host | Clontech |
| E. faecium 8E9 | Ampicillin/vancomycin/linezolid resistant hospital isolate | UMN1 collection |
| E. coli O157:H7 472 | Common pathogenic species | UMN1 collection |
| S. Enteritidis Mh91989 | Chicken isolate; common pathogenic species | UMN1 collection |
| S. 4,[5],12:i:- Mh06225 | Chicken isolate; common pathogenic species | UMN1 collection |
| Plasmid | Description | Reference |
| pHK22 | pACYC184 derivative containing 9.1-kb MccV production fragment | [18] |
| pHK22Δ | pHK22 derivative with mutated cvaC gene | This study |
| pMPES | pHK22Δ derivative containing the ProTeOn+ promoter | This study |
| pMPES:V, L, VspL, N, VspN, VspA, LspA, A, VspH, VspP, VspB | See Table 2 | This study |
| pMPESΔ | pMPES digested with XmaI to eliminate cvaA and cvaB expression | This study |
| pMPESΔ:V | pMPESΔ with V and MccV immunity gene | This study |
| pMPESΔ:L | pMPESΔ with L and MccL immunity gene | This study |
| pMPESΔ:VA | pMPESΔ with VspA and EntA immunity gene | This study |
| pMPESb | pMPES with alternative ribosomal binding site | This study |
| pMPESb:VspA | pMPESb with VspA | This study |
| pMPES_b:VspA_L | pMPESb with VspA and L with their immunity genes | This study |
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Geldart, K.; Forkus, B.; McChesney, E.; McCue, M.; Kaznessis, Y.N. pMPES: A Modular Peptide Expression System for the Delivery of Antimicrobial Peptides to the Site of Gastrointestinal Infections Using Probiotics. Pharmaceuticals 2016, 9, 60. https://doi.org/10.3390/ph9040060
Geldart K, Forkus B, McChesney E, McCue M, Kaznessis YN. pMPES: A Modular Peptide Expression System for the Delivery of Antimicrobial Peptides to the Site of Gastrointestinal Infections Using Probiotics. Pharmaceuticals. 2016; 9(4):60. https://doi.org/10.3390/ph9040060
Chicago/Turabian StyleGeldart, Kathryn, Brittany Forkus, Evelyn McChesney, Madeline McCue, and Yiannis N. Kaznessis. 2016. "pMPES: A Modular Peptide Expression System for the Delivery of Antimicrobial Peptides to the Site of Gastrointestinal Infections Using Probiotics" Pharmaceuticals 9, no. 4: 60. https://doi.org/10.3390/ph9040060




