Metabolomics as a Prospective Tool for Soybean (Glycine max) Crop Improvement
Abstract
:1. Introduction
Relevance as a Multifunctional Crop
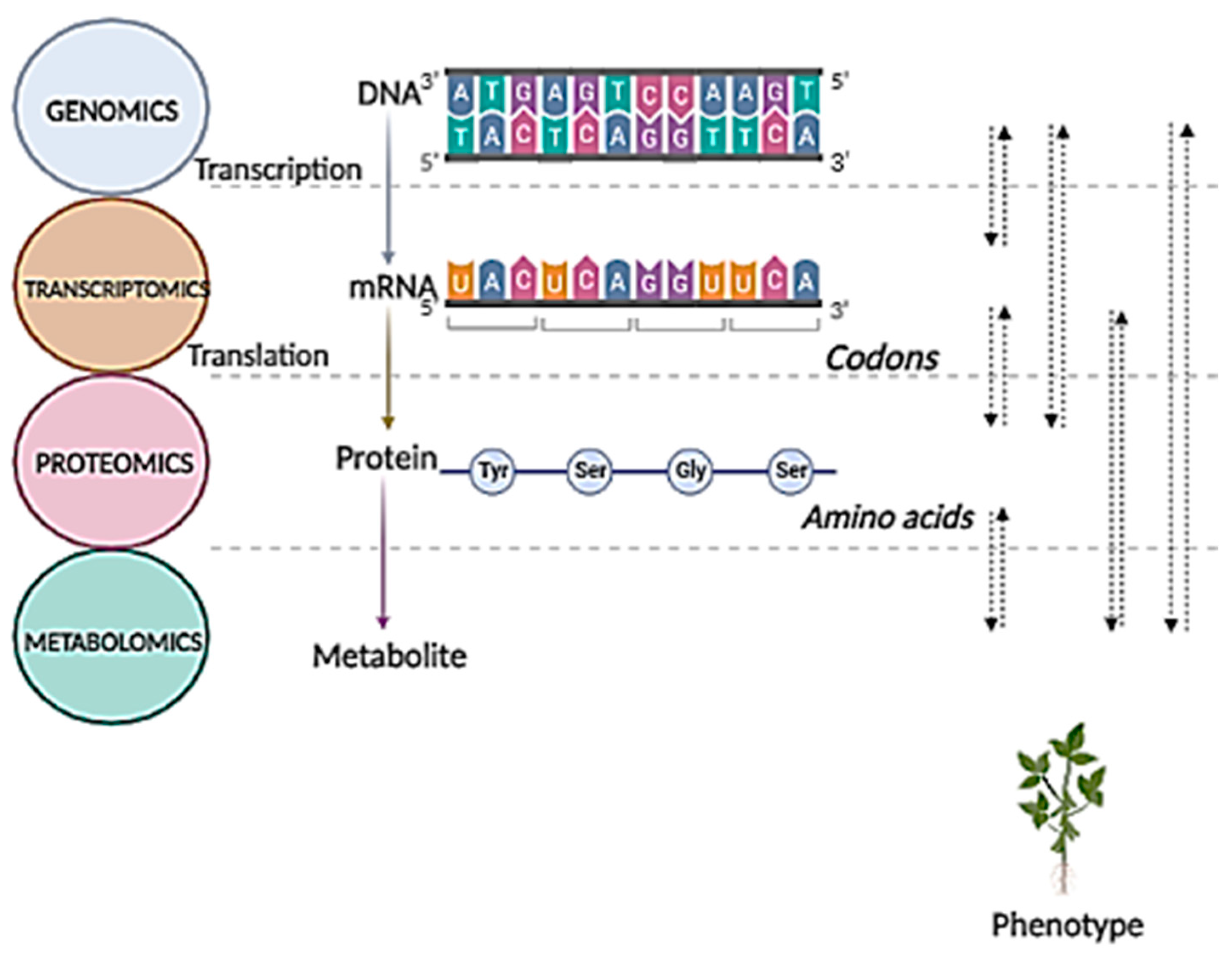
2. Metabolomics at the Forefront of Functional Genomic Approaches
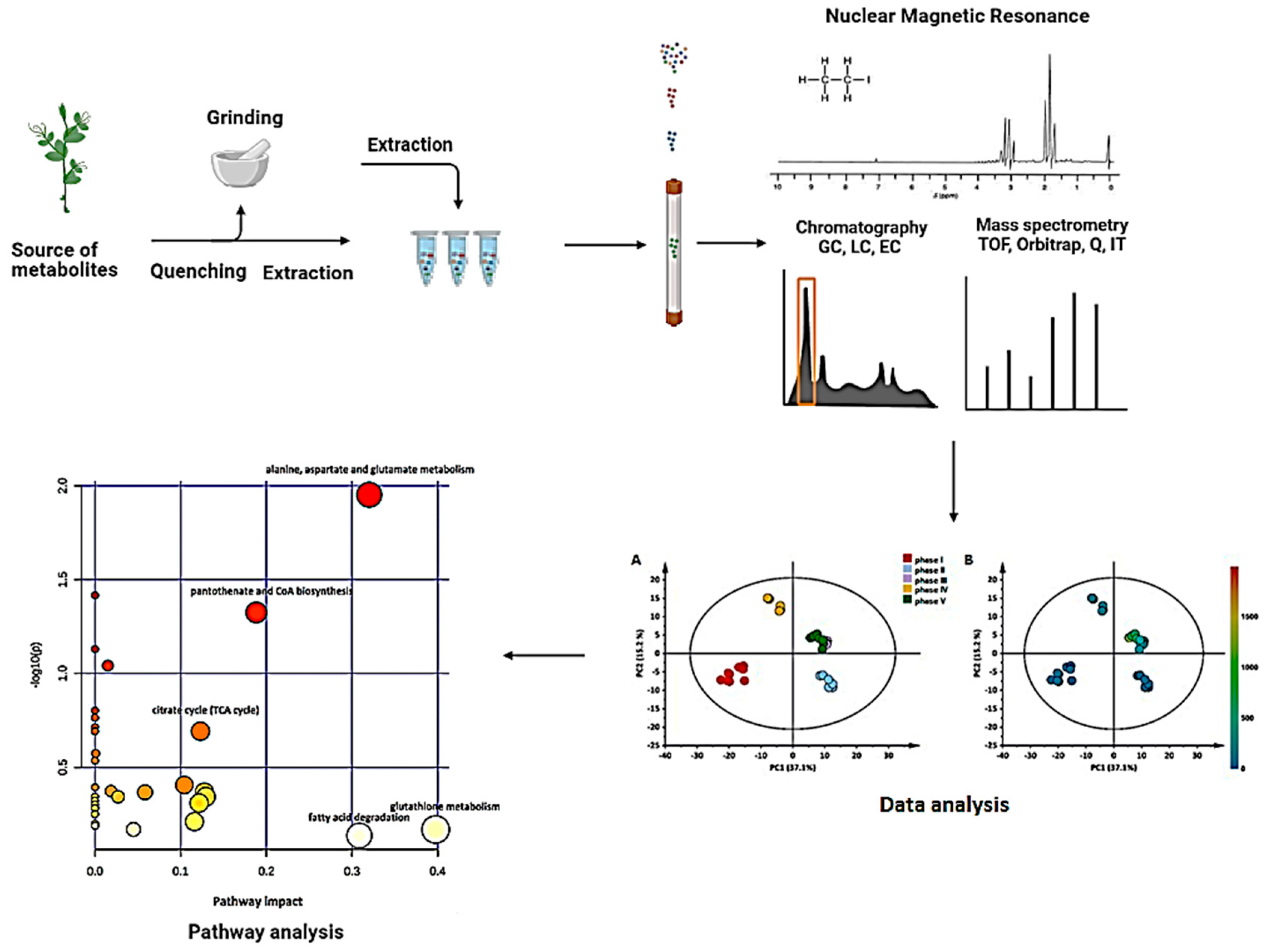
2.1. Sample Preparation
2.2. Data Acquisition
2.3. Data Analysis
2.3.1. Data Visualization (Pre-Processing and Pre-Treatment)
2.3.2. Statistical Modelling
2.4. Metabolite Annotation, Pathway Mapping, Network Correlation and Biological Interpretation
3. Application of Metabolomics as a Prospective Tool to Improve Soybean
4. Concluding Remarks and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sato, D.; Akashi, H.; Sugimoto, M.; Tomita, M. Metabolomic profiling of the response of susceptible and resistant soybean strains to foxglove aphid, Aulacorthum solani Kaltenbach. J. Chromatogr. B 2013, 925, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Ding, C.; Li, W.; Wang, D.; Cui, D. Applications of Metabolomics in the Research of Soybean Plant under Abiotic Stress; Elsevier Ltd.: Amsterdam, The Netherlands, 2019; ISBN 8657188982820. [Google Scholar]
- Yang, D.; Zhang, J.; Ming-xia, L.; Shi, L. Metabolomics analysis reveals the salt-tolerant mechanism in Glycine soja. J. Plant Growth Regul. 2017, 36, 460–471. [Google Scholar] [CrossRef]
- Muhammad, H.; Sumera, A.K.; Abdul, L.K.; Jae-Ho, S.; Bashir, A.; Shin, D.-H.; Lee, I.-J. Exogenous gibberellic acid reprograms soybean to higher growth and salt stress tolerance. J. Agric. Food Chem. 2010, 58, 7226–7232. [Google Scholar] [CrossRef]
- Shinano, T.; Cheng, W.; Saito, K.; Oikawa, A. Metabolomic analysis of night-released soybean root exudates under high- and low-K conditions. Plant Soil 2020, 456, 259–276. [Google Scholar]
- Oliveira, M.C.; Osipitan, O.A.; Begcy, K.; Werle, R. Cover crops, hormones and herbicides: Priming an integrated weed management strategy. Plant Sci. 2020, 301, 110550. [Google Scholar] [CrossRef]
- Razzaq, A.; Wishart, D.S.; Wani, S.H.; Hameed, M.K.; Mubin, M.; Saleem, F. Advances in metabolomics-driven diagnostic breeding and crop improvement. Metabolites 2022, 12, 511. [Google Scholar] [CrossRef]
- Clarke, J.D.; Alexander, D.C.; Ward, D.P.; Ryals, J.A.; Mitchell, M.W.; Wulff, J.E.; Guo, L. Assessment of genetically modified soybean in relation to natural variation in the soybean seed metabolome. Sci. Rep. 2013, 3, 3082. [Google Scholar] [CrossRef]
- García-Villalba, R.; León, C.; Dinelli, G.; Segura-Carretero, A.; Fernández-Gutiérrez, A.; Garcia-Cañas, V.; Cifuentes, A. Comparative metabolomic study of transgenic versus conventional soybean using capillary electrophoresis-time-of-flight mass spectrometry. J. Chromatogr. A 2008, 1195, 164–173. [Google Scholar] [CrossRef]
- Padgette, S.R.; Re, D.B.; Barry, G.F.; Eichholtz, D.E.; Xavier, D.; Fuchs, R.L.; Kishore, G.M.; Fraley, R.T. New weed control opportunities: Development of soybeans with a Roundup Ready™ gene. In Herbicide–Resistant Crops; CRC Press: Boca Raton, FL, USA, 2018; pp. 53–84. [Google Scholar]
- Alberto, C.; Manuel, J.; Oliveira, L.; Mui, S.; Antunes, R. Physiological effects of glyphosate over amino acid profile in conventional and transgenic soybean (Glycine max). Pestic. Biochem. Physiol. 2012, 102, 134–141. [Google Scholar] [CrossRef]
- Lundry, D.R.; Ridley, W.P.; Meyer, J.J.; Riordan, S.G.; Nemeth, M.A.; Trujillo, W.A.; Breeze, M.L.; Sorbet, R. Composition of Grain, forage, and processed fractions from second-generation glyphosate-tolerant soybean, MON 89788, Is equivalent to that of conventional soybean (Glycine max L.). J. Agric. Food Chem. 2008, 2006, 4611–4622. [Google Scholar] [CrossRef]
- Chaudhary, J.; Deshmukh, R.; Mir, Z.A.; Bhat, J.A. Metabolomics: An Emerging Technology for Soybean Improvement; Springer International Publishing: Berlin/Heidelberg, Germany, 2019; ISBN 9783319923994. [Google Scholar]
- Krishnan, H.B.; Song, B.; Oehrle, N.W.; Cameron, J.C.; Jez, J.M. Impact of overexpression of cytosolic isoform of O-acetylserine sulfhydrylase on soybean nodulation and nodule metabolome. Sci. Rep. 2018, 8, 2367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hyung, H.S.; Ryu, W. Metabolomics investigation of flavonoid synthesis in soybean leaves depending on the growth stage. Metabolomics 2014, 10, 833–841. [Google Scholar] [CrossRef]
- Seo, H.S.; Lee, S.; Singh, D.; Shin, H.W.; Cho, S.A.; Lee, C.H. Untargeted metabolite profiling for koji-fermentative bioprocess unravels the effects of varying substrate types and microbial inocula. Food Chem. 2018, 266, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Nahar, K.; Hasanuzzaman, M.; Fujita, M. Heat Stress Responses and Thermotolerance in Soybean; Elsevier Inc.: Amsterdam, The Netherlands, 2016; ISBN 9780128015360. [Google Scholar]
- Gu, E.; Wook, D.; Jang, G.; Hwa, S.; Lee, J.; Bong, S.; Kim, B.; Cho, Y.; Lee, H.; Kim, H. Mass-based metabolomic analysis of soybean sprouts during germination. Food Chem. 2017, 217, 311–319. [Google Scholar] [CrossRef]
- Kwon, Y.S.; Lee, S.; Lee, S.H.; Kim, H.J.; Lee, C.H. Comparative evaluation of six traditional fermented soybean products in East Asia: A metabolomics approach. Metabolites 2019, 9, 183. [Google Scholar] [CrossRef]
- Yun, D.; Kang, Y.; Yun, B.; Kim, E.; Kim, M.; Park, J.S.; Lee, J.; Hong, Y.S. Distinctive metabolism of flavonoid between cultivated and semi-wild soybean unveiled through metabolomics approach. J. Agric. Food Chem. 2016, 64, 5773–5783. [Google Scholar] [CrossRef]
- Bueno, P.C.P.; Lopes, N.P. Metabolomics to characterize adaptive and signaling responses in legume crops under abiotic stresses. ACS Omega 2020, 5, 1752–1763. [Google Scholar] [CrossRef]
- Demers, L.C. Comparative functional genomics characterization of low phytic acid soybeans and virus resistant soybeans. Ph.D. Dissertation, Virginia Polytechnic Institute and State University, Blacksburg, VA, USA, 2020. [Google Scholar]
- Schmidt, M.A.; Herman, E.M. Characterization and functional biology of the soybean aleurone layer. BMC Plant Biol. 2018, 8, 354. [Google Scholar] [CrossRef]
- Yi, L.; Dong, N.; Yun, Y.; Deng, B.; Liu, S.; Zhang, Y.; Liang, Y. Recent advances in chemometric methods for plant metabolomics: A review. Biotechnol. Adv. 2014, 914, 17–34. [Google Scholar] [CrossRef]
- Zampieri, M.; Sekar, K.; Zamboni, N.; Sauer, U. Frontiers of high-throughput metabolomics. Curr. Opin. Chem. Biol. 2017, 36, 15–23. [Google Scholar] [CrossRef]
- Maruyama, Y.; Toya, Y.; Kurokawa, H.; Fukano, Y.; Sato, A.; Umemura, H.; Yamada, K.; Iwasaki, H.; Tobori, N.; Shimizu, H. Characterization of oil-producing yeast Lipomyces starkeyi on glycerol carbon source based on metabolomics and 13C-labeling. Appl. Microbiol. Biotechnol. 2018, 102, 8909–8920. [Google Scholar] [CrossRef]
- Tugizimana, F.; Piater, L.A.; Dubery, I.A. Plant metabolomics: A new frontier in phytochemical analysis. S. Afr. J. Sci. 2013, 109, 18–20. [Google Scholar] [CrossRef]
- Hall, R.D. Plant metabolomics: From holistic hope, to hype, to hot topic. New Phytol. 2006, 169, 453–468. [Google Scholar] [CrossRef] [PubMed]
- Schauer, N.; Fernie, A. Plant metabolomics: Towards biological function and mechanism. Trends Plant Sci. 2006, 11, 508–516. [Google Scholar] [CrossRef] [PubMed]
- Cook, D.W.; Rutan, S.C. Chemometrics for the analysis of chromatographic data in metabolomics investigations. J. Chemom. 2014, 28, 681–687. [Google Scholar] [CrossRef]
- Djande, C.Y.H.; Pretorius, C.; Tugizimana, F.; Piater, L.A.; Dubery, I.A. Metabolomics: A Tool for Cultivar phenotyping and investigation of grain crops. Agronomy 2020, 10, 831. [Google Scholar] [CrossRef]
- Ranjbar, M.R.N.; Zhao, Y.; Tadesse, M.G.; Wang, Y.; Ressom, H.W. Gaussian process regression model for normalization of LC-MS data using scan-level information. Proteome Sci. 2013, 11, S13. [Google Scholar] [CrossRef]
- Kostidis, S.; Addie, R.D.; Morreau, H.; Mayboroda, O.A.; Giera, M. Quantitative NMR analysis of intra- and extracellular metabolism of mammalian cells: A tutorial. Anal. Chim. Acta 2017, 980, 1–24. [Google Scholar] [CrossRef]
- Nandania, J.; Peddinti, G.; Pessia, A.; Kokkonen, M.; Velagapudi, V. Validation and automation of a high-throughput multitargeted method for semiquantification of endogenous metabolites from different biological matrices using tandem mass spectrometry. Metabolites 2018, 8, 44. [Google Scholar] [CrossRef]
- Fuhrer, T.; Zamboni, N. High-throughput discovery metabolomics. Curr. Opin. Biotechnol. 2015, 31, 73–78. [Google Scholar] [CrossRef]
- Niu, Z.; Zhang, W.; Yu, C.; Zhang, J.; Wen, Y. Trends in analytical chemistry recent advances in biological sample preparation methods coupled with chromatography, spectrometry and electrochemistry analysis techniques. Trends Anal. Chem. 2018, 102, 123–146. [Google Scholar] [CrossRef]
- Antignac, J.; Dervilly-pinel, G.; Bizec, B. Le Basics of mass spectrometry based metabolomics. Proteomics 2014, 14, 2369–2388. [Google Scholar] [CrossRef]
- Jiao, L.; Tao, Y.; Wang, W.; Shao, Y.; Mei, L.; Wang, Q.; Dang, J. Preparative isolation of flavonoid glycosides from Sphaerophysa salsula using hydrophilic interaction solid-phase extraction coupled with two-dimensional preparative liquid chromatography. J. Sep. Sci. 2017, 40, 3808–3816. [Google Scholar] [CrossRef] [PubMed]
- Adegbe, A.A.; Larayetan, R.A.; Omojuwa, T.J. Proximate analysis, physicochemical properties and chemical constituents characterization of Moringa oleifera (Moringaceae) seed oil using GC-MS Analysis. Am. J. Chem. 2016, 6, 23–28. [Google Scholar] [CrossRef]
- Khoza, B.S.; Chimuka, L.; Mukwevho, E.; Steenkamp, P.A.; Madala, N.E. The effect of temperature on pressurised hot water extraction of pharmacologically important metabolites as analysed by UPLC-qTOF-MS and PCA. Evidence-based Complement. Altern. Med. 2014, 2014, 1–9. [Google Scholar] [CrossRef]
- Mohammed, N.K.; Yazid, M.; Manap, A.; Tan, C.P.; Muhialdin, B.J.; Alhelli, A.M.; Shobirin, A.; Hussin, M. The Effects of different extraction methods on antioxidant properties, chemical composition, and thermal behavior of black seed (Nigella sativa L.) oil. Evid. -Based Complementary Altern. Med. 2016, 2016, 6273817. [Google Scholar] [CrossRef]
- Bragagnolo, F.S.; Funari, C.S.; Ibáñez, E.; Cifuentes, A. Metabolomics as a tool to study underused soy parts: In search of bioactive compounds. Foods 2021, 10, 1308. [Google Scholar] [CrossRef]
- Louie, K.B.; Kosina, S.M.; Hu, Y.; Otani, H.; de Raad, M.; Kuftin, A.N.; Mouncey, N.J.; Bowen, B.P.; Northen, T.R. Mass spectrometry for natural product discovery. In Comprehensive Natural Products III; Elsevier Ltd.: Amsterdam, The Netherlands, 2020; pp. 263–306. ISBN 9780124095472. [Google Scholar]
- Buszewski, B.; Rafińska, K.; Cvetanović, A.; Walczak, J. Phytochemical analysis and biological activity of Lupinus luteus seeds extracts obtained by supercritical fluid extraction. Phytochem. Lett. 2019, 30, 338–348. [Google Scholar] [CrossRef]
- Teo, C.C.; Tan, S.N.; Yong, J.W.H.; Hew, C.S.; Ong, E.S. Pressurized hot water extraction (PHWE). J. Chromatogr. A 2010, 1217, 2484–2494. [Google Scholar] [CrossRef]
- Dettmer, K.; Aronov, P.A.; Hammock, B.D. Mass spectrometry-based metabolomics. Mass Spectrom. Rev. 2007, 26, 51–78. [Google Scholar] [CrossRef]
- Porzel, A.; Farag, M.A.; Mülbradt, J.; Wessjohann, L.A. Metabolite profiling and fingerprinting of Hypericum species: A comparison of MS and NMR metabolomics. Metabolomics 2014, 10, 574–588. [Google Scholar] [CrossRef]
- Sehlakgwe, P.F.; Lall, N.; Prinsloo, G. 1H-NMR Metabolomics and LC-MS Analysis to determine seasonal variation in a cosmeceutical plant Leucosidea sericea. Front. Pharmacol. 2020, 11, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Allwood, J.W.; Goodacre, R. An introduction to liquid chromatography—mass spectrometry instrumentation applied in plant metabolomic analyses. Phytochem. Anal. 2010, 21, 33–47. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.-J.; Schultz, A.W.; Wang, J.; Johnson, C.H.; Yannone, S.M.; Patti, G.J.; Siuzdak, G. Liquid chromatography quadrupole time-of-flight mass spectrometry characterization of metabolites guided by the METLIN database. Nat. Protoc. 2013, 8, 451–460. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.H.; Ivanisevic, J.; Siuzdak, G. Metabolomics: Beyond biomarkers and towards mechanisms. Nat. Rev. Mol. Cell Biol. 2016, 17, 451–459. [Google Scholar] [CrossRef]
- Mahrous, E.A.; Farag, M.A. Two dimensional NMR spectroscopic approaches for exploring plant metabolome: A review. J. Adv. Res. 2015, 6, 3–15. [Google Scholar] [CrossRef]
- Kuhn, S.; Colreavy-donnelly, S. An integrated approach for mixture analysis using MS and NMR techniques. Faraday Discuss 2019, 218, 339–353. [Google Scholar] [CrossRef]
- Tugizimana, F.; Ncube, E.N.; Steenkamp, P.A.; Dubery, I.A. Metabolomics-derived insights into the manipulation of terpenoid synthesis in Centella asiatica cells by methyl jasmonate. Plant Biotechnol. Rep. 2015, 9, 125–136. [Google Scholar] [CrossRef]
- Hantao, L.W.; de Lima Ribeiro, F.A.; Passador, M.M.; Furtado, E.L.; Poppi, R.J.; Gozzo, F.C.; Augusto, F. Metabolic profiling by ultra-performance liquid chromatography-mass spectrometry and parallel factor analysis for the determination of disease biomarkers in Eucalyptus. Metabolomics 2014, 10, 1318–1325. [Google Scholar] [CrossRef]
- Putri, S.P.; Yamamoto, S.; Tsugawa, H.; Fukusaki, E. Current metabolomics. Technological advances. J. Biosci. Bioeng. 2013, 116, 9–16. [Google Scholar] [CrossRef]
- Ernst, M.; Silva, D.B.; Silva, R.R.; Vêncio, R.Z.N.; Lopes, N.P. Mass spectrometry in plant metabolomics strategies: From analytical platforms to data acquisition and processing. Nat. Prod. Rep. 2014, 31, 784. [Google Scholar] [CrossRef] [PubMed]
- Gika, H.G.; Wilson, I.D.; Theodoridis, G.A. The Role of Mass Spectrometry in Nontargeted Metabolomic, 1st ed.; Elsevier: Amsterdam, The Netherlands, 2014; Volume 63, ISBN 0166526X. [Google Scholar]
- Wolfender, J.-L.; Aurelien, G.M.; Bertrand, T.S. Current approaches and challenges for the metabolite profiling of complex natural extracts. J. Chromatogr. A 2015, 1382, 136–164. [Google Scholar] [CrossRef] [PubMed]
- Van Den Berg, R.A.; Hoefsloot, H.C.J.; Westerhuis, J.A.; Smilde, A.K.; Van DerWerf, M.J. Centering, scaling, and transformations: Improving the biological information content of metabolomics data. BMC Genomics 2006, 7, 142. [Google Scholar] [CrossRef] [PubMed]
- Castillo, S.; Gopalacharyulu, P.; Yetukuri, L.; Oresic, M. Algorithms and tools for the preprocessing of LC—MS metabolomics data. Chemom. Intell. Lab. Syst. 2011, 108, 23–32. [Google Scholar] [CrossRef]
- Xu, Y.F.; Lu, W.; Rabinowitz, J.D. Avoiding misannotation of in-source fragmentation products as cellular metabolites in liquid chromatography-mass spectrometry-based metabolomics. Anal. Chem. 2015, 87, 2273–2281. [Google Scholar] [CrossRef]
- Allwood, J.W.; De Vos, R.C.H.; Moing, A.; Deborde, C.; Erban, A.; Kopka, J.; Goodacre, R.; Hall, R.D. Plant metabolomics and its potential for systems biology research. In Methods in Systems Biology; Elsevier Inc.: Amsterdam, The Netherlands, 2011; Volume 500, pp. 299–336. ISBN 9780123851185. [Google Scholar]
- Trygg, J.; Holmes, E.; Lundstedt, T. Chemometrics in metabonomics. J. Proteome Res. 2007, 6, 469–479. [Google Scholar] [CrossRef]
- Allwood, J.W.; Ellis, D.I.; Goodacre, R. Metabolomic technologies and their application to the study of plants and plant–host interactions. Physiol. Plant. 2008, 132, 117–135. [Google Scholar] [CrossRef]
- Saccenti, E.; Hoefsloot, H.C.J.; Smilde, A.K.; Westerhuis, J.A.; Hendriks, M.M.W.B. Reflections on univariate and multivariate analysis of metabolomics data. Metabolomics 2014, 10, 361–374. [Google Scholar] [CrossRef]
- Olivon, F.; Roussi, F.; Litaudon, M.; Touboul, D. Optimized experimental workflow for tandem mass spectrometry molecular networking in metabolomics. Anal. Bioanal. Chem. 2017, 409, 5767–5778. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Furumichi, M.; Morishima, K.; Tanabe, M. New approach for understanding genome variations in KEGG. Nucleic Acids Res. 2019, 47, D590–D595. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y. KEGG Mapper for inferring cellular functions from protein sequences. Protein Sci. 2020, 29, 28–35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Hu, B.; Liu, W.; Qin, W.; Wu, H.; Zhang, J.; Yang, C.; Deng, J.; Shu, K.; Du, J.; et al. Metabolomic tool to identify soybean [Glycine max (L.) Merrill] germplasms with a high level of shade tolerance at the seedling stage. Sci. Rep. 2017, 7, 42478. [Google Scholar] [CrossRef] [PubMed]
- Fukushima, A.; Kusano, M. Recent Progress in the development of metabolome databases for plant systems biology. Front. Plant Sci. 2013, 4, 73. [Google Scholar] [CrossRef]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.M.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed minimum reporting standards for chemical analysis: Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef]
- Bhalla, R.; Narasimhan, K.; Swarup, S. Metabolomics and its role in understanding cellular responses in plants. Plant Cell Rep. 2005, 24, 562–571. [Google Scholar] [CrossRef]
- Maruyama, K.; Urano, K.; Kusano, M.; Sakurai, T.; Takasaki, H.; Kishimoto, M. Metabolite/phytohormone—gene regulatory networks in soybean organs under dehydration conditions revealed by integration analysis. Plant J. 2020, 103, 197–211. [Google Scholar] [CrossRef]
- Silvente, S.; Sobolev, A.P.; Lara, M. Metabolite adjustments in drought tolerant and sensitive soybean genotypes in response to water stress. PLoS ONE 2012, 7, e38554. [Google Scholar] [CrossRef]
- Wang, X.; Guo, R.; Li, M.; Liu, Y.; Zhao, M.; Fu, H.; Liu, X.; Wang, S. Metabolomics reveals the drought - tolerance mechanism in wild soybean (Glycine soja). Acta Physiol. Plant. 2019, 41, 1–11. [Google Scholar] [CrossRef]
- Díaz-cruz, G.A.; Cassone, B.J. A tale of survival: Molecular defense mechanisms of soybean to overcome soybean mosaic virus infection physiological and molecular plant pathology. A tale of survival: Molecular defense mechanisms of soybean to overcome Soybean Mosaic Virus infection. Physiol. Mol. Plant Pathol. 2017, 102, 79–87. [Google Scholar] [CrossRef]
- Hu, B.-Y.; Yang, C.-Q.; Iqbal, N.; Deng, J.-C.; Zhang, J.; Yang, W.-Y.; Liu, J. Development and validation of a GC–MS method for soybean organ-specific metabolomics. Plant Prod. Sci. 2018, 21, 215–224. [Google Scholar] [CrossRef]
- Coutinho, I.D.; Henning, L.M.M.; Döpp, S.A.; Nepomuceno, A.; Moraes, L.A.C.; Marcolino-Gomes, J.; Richter, C.; Schwalbe, H.; Colnago, L.A. Flooded soybean metabolomic analysis reveals important primary and secondary metabolites involved in the hypoxia stress response and tolerance. Environ. Exp. Bot. 2018, 153, 176–187. [Google Scholar] [CrossRef] [Green Version]
- Kang, W.; Chen, L.; Wang, Y.; Zhu, X.; Liu, X.; Duan, Y. Bacillus simplex treatment promotes soybean defence against soybean cyst nematodes: A metabolomics study using GC-MS. PLoS ONE 2020, 15, e0237194. [Google Scholar] [CrossRef] [PubMed]
- Ahsan, N.; Nakamura, T.; Komatsu, S. Differential responses of microsomal proteins and metabolites in two contrasting cadmium (Cd -accumulating soybean cultivars under Cd stress. Amino Acids 2012, 42, 317–327. [Google Scholar] [CrossRef]
- Rabara, R.C.; Tripathi, P.; Rushton, P.J. Comparative metabolome profile between tobacco and soybean grown under water-stressed conditions. Biomed Res. Int. 2017, 2017, 3065251. [Google Scholar] [CrossRef]
- Das, A.; Rushton, P.J.; Rohila, J.S. Metabolomic profiling of soybeans (Glycine max L.) Reveals the Importance of Sugar and Nitrogen. Plants 2017, 6, 21. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Hu, C.; Hussain, S.; Tan, Q.; Wu, S. Ecotoxicology and environmental safety metabolomics analysis reveals potential mechanisms of tolerance to excess molybdenum in soybean seedlings. Ecotoxicol. Environ. Saf. 2018, 164, 589–596. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Xu, J.; Wang, X.; Fu, H.; Zhao, M.; Wang, H.; Shi, L. Photosynthetic characteristics and metabolic analyses of two soybean genotypes revealed adaptive strategies to low-nitrogen stress. J. Plant Physiol. 2018, 229, 132–141. [Google Scholar] [CrossRef]
- Jiao, Y.; Bai, Z.; Xu, J.; Zhao, M.; Khan, Y.; Hu, Y.; Shi, L. Metabolomics and its physiological regulation process reveal the salt- tolerant mechanism in Glycine soja seedling roots. Plant Physiol. Biochem. 2018, 126, 187–196. [Google Scholar] [CrossRef]
- Li, M.; Guo, R.; Jiao, Y.; Jin, X.; Zhang, H.; Shi, L. Comparison of salt tolerance in Soja Based on metabolomics of seedling roots. Front. Plant Sci. 2017, 8, 1101. [Google Scholar] [CrossRef]
- Xu, G.; Singh, S.; Barnaby, J.; Buyer, J.; Reddy, V.; Sicher, R. Effects of growth temperature and carbon dioxide enrichment on soybean seed components at different stages of development. Plant Physiol. Biochem. 2016, 108, 313–322. [Google Scholar] [CrossRef]
- Copley, T.R.; Aliferis, K.A.; Kliebenstein, D.J.; Jabaji, S.H. An integrated RNAseq- 1 H NMR metabolomics approach to understand soybean primary metabolism regulation in response to Rhizoctonia foliar blight disease. BMC Plant Biol. 2017, 17, 84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hrbek, V.; Krtkova, V.; Rubert, J.; Chmelarova, H.; Demnerova, K.; Ovesna, J.; Hajslova, J. Metabolomic strategies based on High-Resolution Mass Spectrometry as a tool for recognition of GMO (MON 89788 Variety) and Non-GMO Soybean: A critical assessment of two complementary methods. Food Anal. Methods 2017, 10, 3723–3737. [Google Scholar] [CrossRef]
- Fu, H.; Guo, R.; Shen, W.Y.; Li, M.X.; Liu, Y.; Zhao, M.L.; Wang, X.X.; Liu, X.Y. Changes in the metabolome of two soybean genotypes under drought stress. Russ. J. Plant Physiol. 2020, 67, 472–481. [Google Scholar] [CrossRef]
- Yang, A.; Kong, L.; Wang, H.; Yao, X.; Xie, F.; Wang, H. Response of soybean root to phosphorus deficiency under sucrose feeding: Insight from morphological and metabolome characterizations. BioMed Res. Int. 2020, 2020, 2148032. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Xu, J.; Guo, R.; Liu, Y.; Wang, S.; Wang, H.; Ullah, A.; Shi, L. Identifying the metabolomics and physiological differences among Soja in the early flowering stage. Plant Physiol. Biochem. 2019, 139, 82–91. [Google Scholar] [CrossRef]
- Salloum, M.S.; Insani, M.; Monteoliva, M.I.; Menduni, M.F.; Silvente, S.; Carrari, F.; Luna, C. Metabolic responses to arbuscular mycorrhizal fungi are shifted in roots of contrasting soybean genotypes. Mycorrhiza 2019, 29, 459–473. [Google Scholar] [CrossRef] [PubMed]
- Pi, E.; Xu, J.; Li, H.; Fan, W.; Zhu, C.; Zhang, T.; Jiang, J.; He, L.; Lu, H.; Wang, H.; et al. Enhanced salt tolerance of rhizobia-inoculated soybean correlates with decreased phosphorylation of the transcription factor GmMYB183 and altered flavonoid biosynthesis enhanced salt tolerance of rhizobia- inoculated soybean correlates with decreased phos. Mol. Cell. Proteom. 2019, 18, 2225–2243. [Google Scholar] [CrossRef]
- John, K.M.M.; Khan, F.; Luthria, D.L.; Matthews, B.; Garrett, W.M.; Natarajan, S. Proteomic and metabolomic analysis of minimax and Williams 82 soybeans grown under two different conditions. J. Food Biochem. 2017, 41, e12404. [Google Scholar] [CrossRef]
- Pastor, V.; Luna, E.; Ton, J.; Flors, V. Primed plants do not forget. Environ. Exp. Bot. 2012, 94, 46–56. [Google Scholar] [CrossRef]
- Chen, W.; Gao, Y.; Xie, W.; Gong, L.; Lu, K.; Wang, W.; Li, Y.; Liu, X. Genome-wide association analyses provide genetic and biochemical insights into natural variation in rice metabolism. Nat. Publ. Gr. 2014, 46, 714–721. [Google Scholar] [CrossRef]
- Yadav, C.B.; Srivastava, R.K.; Mur, L.A.J.; Yadav, R.S. Metabolite diversity and metabolic genome-wide marker traits in pearl millet grains. Cells 2021, 10, 3076. [Google Scholar] [CrossRef] [PubMed]
| Objective of the Study | Analytical Platform | Tissue | Other Omics | Main Finding | References |
|---|---|---|---|---|---|
| Analyzed the organ specificity of metabolites and identification of the features of their regulatory networks in dehydrated soybeans | GC-TOF-MS LC-MS | Leaves Stems Roots | Transcriptomics | ABA is the most highly dehydration-inducible phytohormone in plant aerial parts. | [74] |
| Investigated metabolite changes in relation to physiological responses two soybean genotypes with varying drought tolerance | 1H NMR | Leaves Nodule | Markers important for determining water stress response were identified. | [75] | |
| Elucidated the mechanism behind drought tolerance in drought-tolerant wild soybean | GC-MS | Leaves | Drought-stress mechanisms include the accumulation of osmotic chemicals, as well as an increase in energy and secondary antioxidant metabolism. Drought resistance in wild soybeans. | [76] | |
| Described the metabolic changes in soybean leaves ten days after Soybean mosaic virus infection (SMV) | LC-MS/MS | Leaves | Transcriptomics | There were significant changes in amino acid concentrations in connection to viral infection at the metabolomic level. | [77] |
| Investigated the potential organ-specific resistance mechanism of soybean to F. Moniliforme | GC–MS | Seeds Pods | F. Moniliforme disrupted amino acid metabolism in soybean seeds, and metabolic pathways involved to energy conversion in soybean pods responded substantially to fungal infection. | [78] | |
| Examining the responses to flooding stress in roots and leaves of two soybean cultivars (BR4 and Embrapa 45, sensitive and moderately tolerant to flooding stress, respectively). | 1H NMR | Roots Leaves | Different reactions were observed in the roots and leaves, as well as in flood-tolerant and flood-sensitive cultivars. The majority of the molecules that have transformed are associated to carbon and nitrogen metabolism, as well as the phenylpropanoid pathway. | [79] | |
| Two wild soybean types with varying salt tolerance were chosen, and metabolic alterations in response to neutral-salt stress and alkali-salt stress were studied. | GC–MS | Leaf | The salt-tolerant wild soybean modifies amino acid and organic acid metabolism to generate more suitable solutes and promote the TCA cycle to produce more ATP. | [3] | |
| Investigated the metabolic changes in soybean cyst nematodes after treatment with Sneb545Bacillus simplex. Roots of SCN-infected soybeans | GC-MS | Root | Soybeans treated with Sneb545 have certain characteristics of SCN disease-resistant soybeans. | [80] | |
| Investigated Cd absorption and translocation in two different Cd-accumulating soybean cultivars | CE-MS | Roots | Proteomics | In the Enrei cultivar under Cd stress, amino acids linked to Cd-chelating pathways are quite active. | [81] |
| Investigated drought tolerance in tobacco and soybean plants to unravel metabolic pathways affected by increasing dehydration | LC-MS LC-MS/MS GC-MS | Root Leaf | In both species, the accumulation of metabolites is strongly linked to the degree of dehydration. | [82] | |
| Profiled leaf metabolites under control conditions, drought, and heat stress in a controlled setting. | LC-MS GC-MS | Leaves | Drought and heat stress were found to affect metabolites for various cellular processes which regulate carbohydrate metabolism, amino acid metabolism, peptide metabolism, and purine and pyrimidine biosynthesis. | [83] | |
| Investigated changes in the metabolic profiles of leaves and roots of soybean (Glycine max L.) Seedlings cultivated under normal and excess Mo conditions. | LC-MS/MS | Roots Leaves | Mo stress induced only lipid metabolism and salicylic acid buildup in leaves, whilst in roots the ascorbate–glutathione metabolism and flavonoid/isoflavone biosynthesis significantly increased. | [84] | |
| Analyzed of two soybean genotypes at the metabolomic level revealed the mechanism of low-nitrogen tolerance. | GC–MS | Leaves Roots | In order to tolerate low nitrogen, wild soybean synthesizes favorable secondary metabolites under low-nitrogen stress. | [85] | |
| Examined metabolomics features of wild soybean under several forms of salt stress to determine salt-tolerant processes in wild soybean in the field | GC–MS | Roots | Under neutral-salt stress, the salt-tolerant wild soybean showed enhanced amino acid, carbohydrate, and polyol metabolisms, whereas under alkali-salt stress, it showed improved organic acid, amino acid, and tricarboxylic acid metabolisms. | [86] | |
| Explored the salt tolerance-related mechanisms among Soja, wild soybean, semi-wild soybean, and cultivated soybean under two types of salt stress | GC–MS | Roots | Carbon and nitrogen metabolism, as well as the tricarboxylic acid (TCA) cycle and receiver operating properties (particularly phenolic substance metabolism) of seedling roots, were critical for salt stress resistance and demonstrated a steady decreasing trend from wild soybean to cultivated soybean. | [87] | |
| Determined the effects of growth temperature and carbon dioxide enrichment on soybean seed components at different stages of development | GC–MS | Seeds | CO2 (enrichment) treatments significantly changed the composition of early seeds but had little effect on mature seeds. Treatment effects on seed constituents were ranked as follows: Age > Temperature > CO2. | [88] | |
| Characterized the resistance of soybeans to foxglove aphid, Aulacorthum solani Kaltenbach, at the metabolite level. | CE–TOF–MS | Leaves | Differences in the amino acids in the soybean leaves influenced the free amino acids found in the aphids, which might be implicated in aphid resistance. | [1] | |
| Investigated variations in soybean metabolism in response to R. solani infection during early and late disease phases, focusing on the regulation of soybean primary metabolism and oxidative stress tolerance | 1H NMR | Leaves | Transcriptomics | In response to R. solani infection, significant changes in soybean primary metabolism occurred and metabolite levels involved in redox reactions and ROS signaling were also recorded. | [89] |
| Distinguished between genetically modified organisms (Monsanto 89,788 variety) and organic soybeans | DART-HRMS HPLC-HMRS | Seeds | The most important markers were found to be phosphatidylcholines and sugars. | [90] | |
| Compared the response mechanisms of wild and cultivated soybean to water stress | GC–MS | Leaves | Drought tolerance mechanisms included increasing primary metabolism to control osmotic potential, synthesizing desirable secondary metabolites and fatty acids, and maintaining a symbiotic relationship. | [91] | |
| Explored global metabolomic modifications in low-P-tolerant (Liaodou, L13) and low-P-sensitive (Tiefeng 3, T3) soybean genotypes | LC-MS | Root | Metabolite profiles of both genotypes differed in their responses as numbers of metabolites were exclusively and differentially regulated within each genotype. | [92] | |
| Examined the impact of overexpressing OASS on soybean nodulation and nodule metabolome | LC-MS GC-MS | Nodules | There is a slight decrease in the availability of energy metabolites to OASS overexpressing soybean nodules, which is then offset by the breakdown of cellular components to meet the nodule energy metabolism needs. | [14] | |
| Evaluated root exudates of two soybean cultivars grown under low-, normal-, and high-K+ conditions | CE–TOF–MS | Root | Soybean cultivars differ in their capacity to release root metabolites by altering the exudation of certain metabolites for improved adaptability to high- and low-K conditions. | [5] | |
| Investigated the cellular metabolism-related differences among salt-tolerant wild soybean (W2), salt- sensitive wild soybean (W1) and cultivated soybean (C) in the early flowering stage to reveal the adaptive mechanisms. | GC–TOF–MS | Leaf | Carbohydrate and organic acid metabolism were relatively greater, while the amino acid content and secondary metabolism level were lower in C than W1 | [93] | |
| Evaluated the metabolic responses of improved (I-1) and unimproved (UI-4) soybean genotypes after AM root colonization | GC-MS | Roots | The I-1 genotype has lower quantities of isoflavonoids and alpha-tocopherol and greater levels of malondialdehyde, that can affect the soybean-AM symbiosis. | [94] | |
| Investigated secondary metabolites produced when soybean plants were infected by A. Besseyi. | LC–ESI–MS–MS | Root | There were metabolome variations in root defensive chemicals in response to A. Besseyi attack, as indicated by an increase in the level of flavonoids. | [68] | |
| Identify metabolic changes in soybean roots treated with rhizobia inoculation and salt | LC–TOFMS | Root | Phosphoproteomics | Rhizobia symbiosis enables the soybean plant to adapt with the negative consequences of high soil salt, mostly by increasing ROS scavenging activities. | [95] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ncube, E.; Mohale, K.; Nogemane, N. Metabolomics as a Prospective Tool for Soybean (Glycine max) Crop Improvement. Curr. Issues Mol. Biol. 2022, 44, 4181-4196. https://doi.org/10.3390/cimb44090287
Ncube E, Mohale K, Nogemane N. Metabolomics as a Prospective Tool for Soybean (Glycine max) Crop Improvement. Current Issues in Molecular Biology. 2022; 44(9):4181-4196. https://doi.org/10.3390/cimb44090287
Chicago/Turabian StyleNcube, Efficient, Keletso Mohale, and Noluyolo Nogemane. 2022. "Metabolomics as a Prospective Tool for Soybean (Glycine max) Crop Improvement" Current Issues in Molecular Biology 44, no. 9: 4181-4196. https://doi.org/10.3390/cimb44090287






