Correlation between the RNA Expression and the DNA Methylation of Estrogen Receptor Genes in Normal and Malignant Human Tissues
Abstract
:1. Introduction
2. Methods
2.1. Data Retrieval and Analysis of RNA Expression of Estrogen Receptors in Normal Tissues
2.2. Comparing the Expression of Estrogen Receptors in Tumor and Normal Tissues
2.3. Analysis of DNA Methylation of Estrogen Receptors and Its Correlation with RNA Levels
2.4. Estimating the Relevance between CG Methylation and Tumor Survival
2.5. Statistical Analysis
3. Results
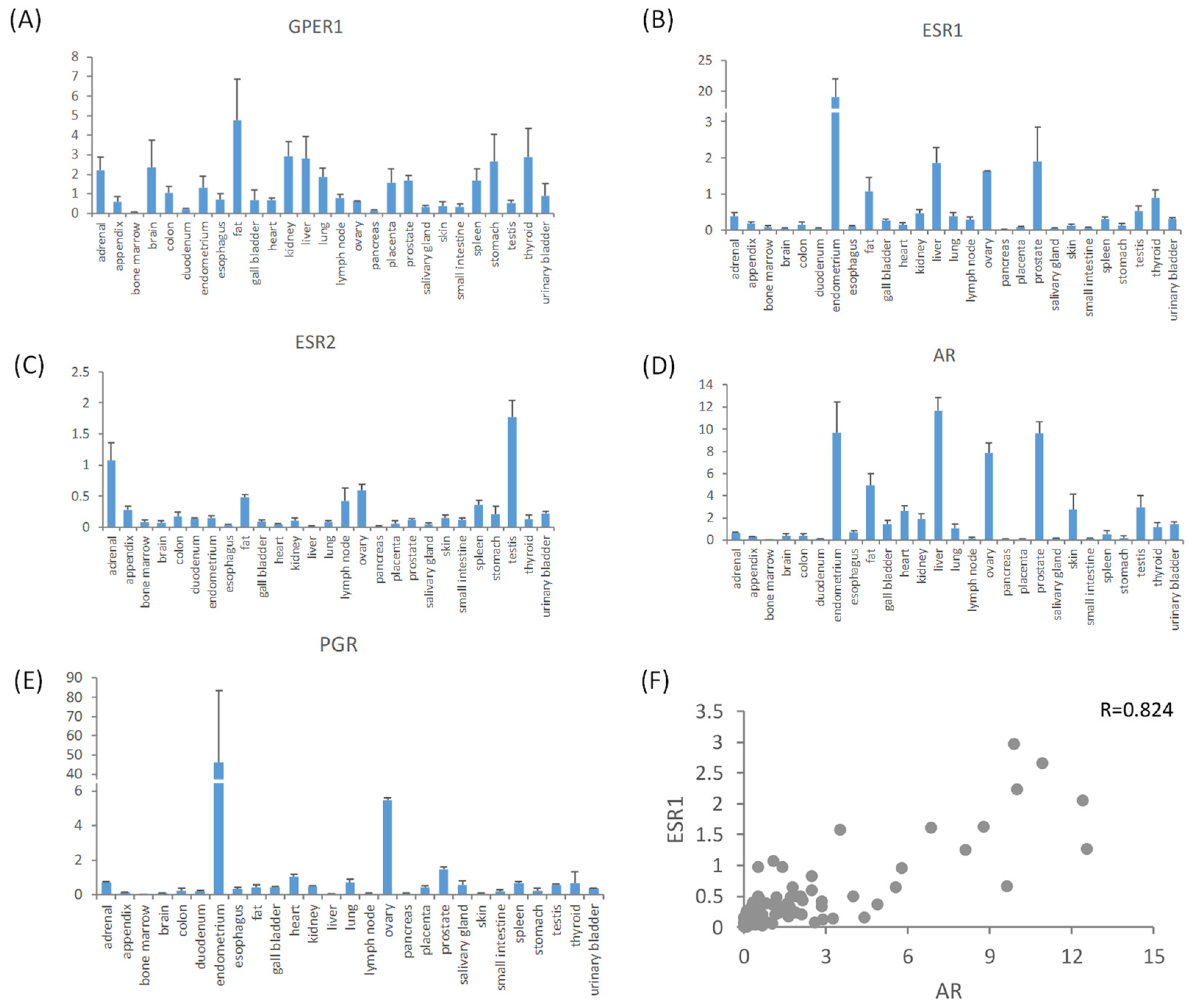
3.1. Expression Profiles of Estrogen Receptors in Normal Tissues
3.2. Expression Features of Estrogen Receptors in Malignant Tissues
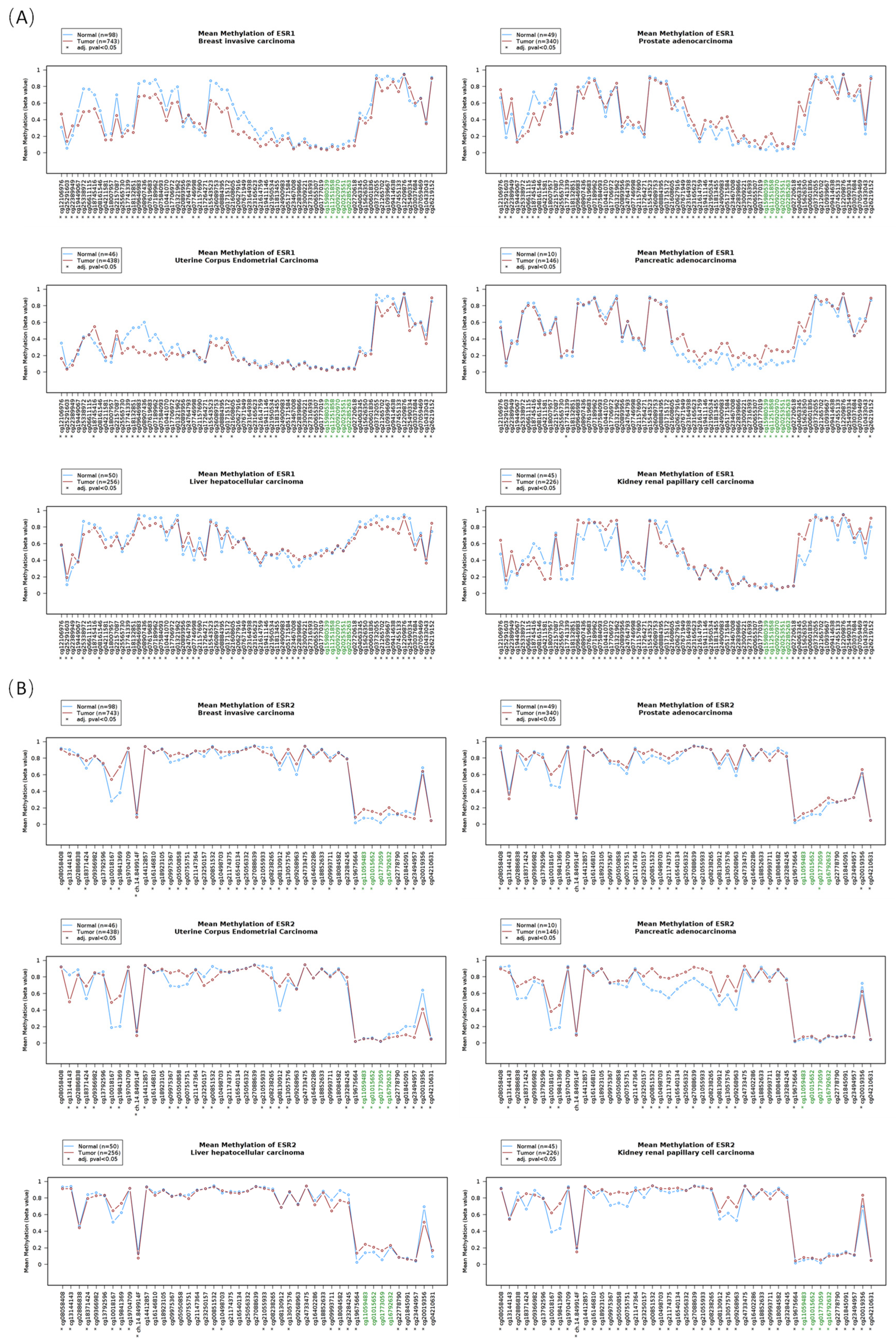
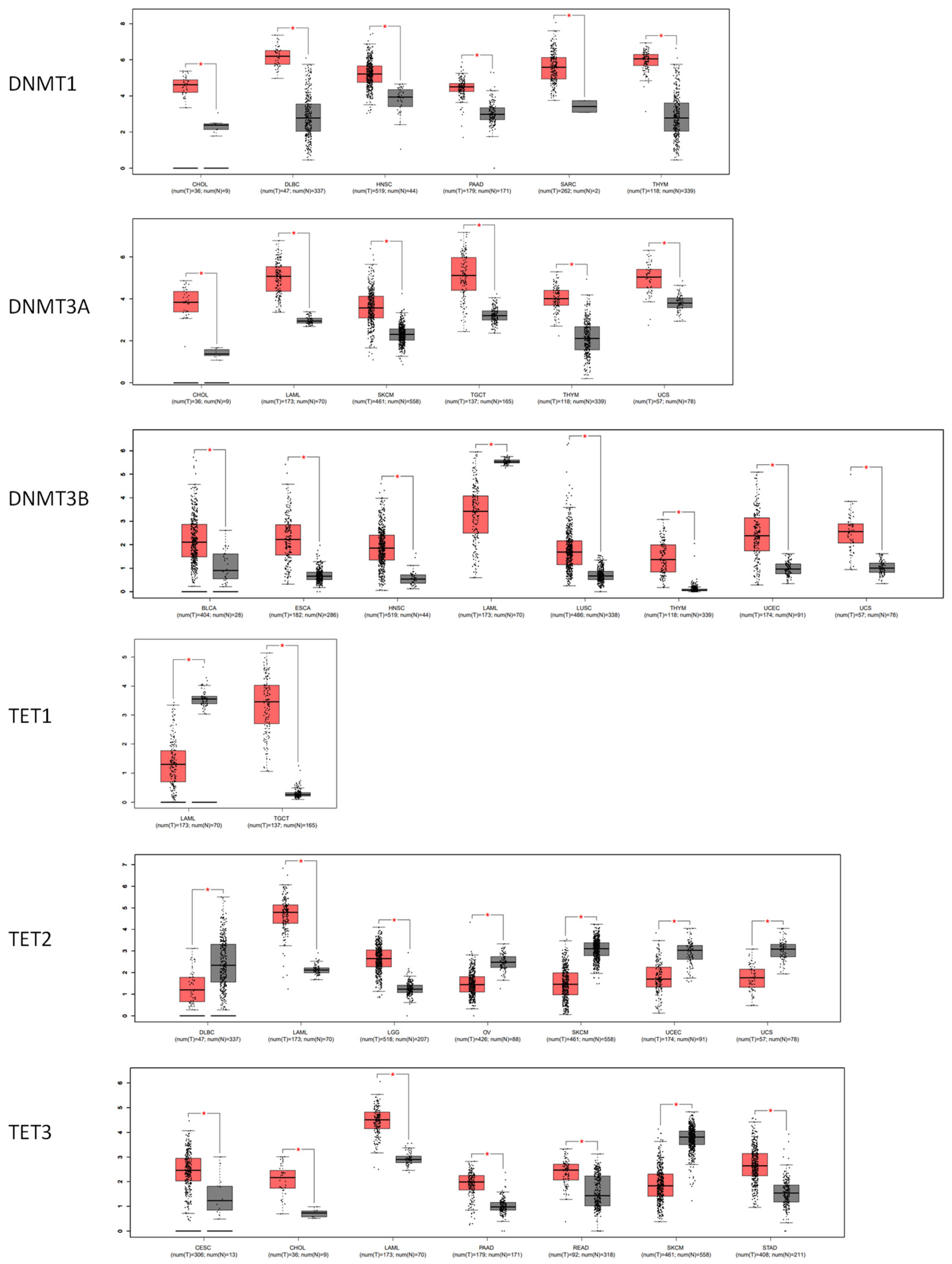
3.3. The DNA Methylation Profile of Estrogen Receptor Genes
3.4. The Correlation between the DNA Methylation and the RNA Level of Estrogen Receptors
3.5. The Correlation between DNA Methylation and Tumor Survival
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Miller, D.D.; Shaw, L. Sex bias in the care of patients with cardiovascular disease? N. Engl. J. Med. 1994, 331, 883. [Google Scholar] [CrossRef] [PubMed]
- Heldring, N.; Pike, A.; Andersson, S.; Matthews, J.; Cheng, G.; Hartman, J.; Tujague, M.; Ström, A.; Treuter, E.; Warner, M.; et al. Estrogen receptors: How do they signal and what are their targets. Physiol. Rev. 2007, 87, 905–931. [Google Scholar] [CrossRef] [PubMed]
- Hannan, F.M.; Elajnaf, T.; Vandenberg, L.N.; Kennedy, S.H.; Thakker, R.V. Hormonal regulation of mammary gland development and lactation. Nat. Rev. Endocrinol. 2023, 19, 46–61. [Google Scholar] [CrossRef] [PubMed]
- Lemaître, J.-F.; Ronget, V.; Tidière, M.; Allainé, D.; Berger, V.; Cohas, A.; Colchero, F.; Conde, D.A.; Garratt, M.; Liker, A.; et al. Sex differences in adult lifespan and aging rates of mortality across wild mammals. Proc. Natl. Acad. Sci. USA 2020, 117, 8546–8553. [Google Scholar] [CrossRef] [PubMed]
- Filardo, E.J.; Quinn, J.A.; Bland, K.I.; Frackelton, A.R., Jr. Estrogen-induced activation of Erk-1 and Erk-2 requires the G protein-coupled receptor homolog, GPR30, and occurs via trans-activation of the epidermal growth factor receptor through release of HB-EGF. Mol. Endocrinol. 2000, 14, 1649–1660. [Google Scholar] [CrossRef]
- Filardo, E.; Quinn, J.; Pang, Y.; Graeber, C.; Shaw, S.; Dong, J.; Thomas, P. Activation of the novel estrogen receptor G protein-coupled receptor 30 (GPR30) at the plasma membrane. Endocrinology 2007, 148, 3236–3245. [Google Scholar] [CrossRef] [PubMed]
- Hirtz, A.; Rech, F.; Dubois-Pot-Schneider, H.; Dumond, H. Estrogen signaling in healthy and tumor brain. Steroids 2023, 199, 109285. [Google Scholar] [CrossRef]
- Lahita, R.G. The role of sex hormones in systemic lupus erythematosus. Curr. Opin. Rheumatol. 1999, 11, 352–356. [Google Scholar] [CrossRef]
- Moulton, V.R. Sex Hormones in Acquired Immunity and Autoimmune Disease. Front. Immunol. 2018, 9, 2279. [Google Scholar] [CrossRef]
- Soldan, S.S.; Retuerto, A.I.A.; Sicotte, N.L.; Voskuhl, R.R. Immune modulation in multiple sclerosis patients treated with the pregnancy hormone estriol. J. Immunol. 2003, 171, 6267–6274. [Google Scholar] [CrossRef]
- Nekrasova, I.; Shirshev, S. Estriol in regulation of cell-mediated immune reactions in multiple sclerosis. J. Neuroimmunol. 2020, 349, 577421. [Google Scholar] [CrossRef] [PubMed]
- Meyer, C.E.; Smith, A.W.; Padilla-Requerey, A.A.; Farkhondeh, V.; Itoh, N.; Itoh, Y.; Gao, J.L.; Herbig, P.D.; Nguyen, Q.; Ngo, K.H.; et al. Neuroprotection in Cerebral Cortex Induced by the Pregnancy Hormone Estriol. Lab. Investig. 2023, 103, 100189. [Google Scholar] [CrossRef] [PubMed]
- Yu, N.Y.-L.; Hallström, B.M.; Fagerberg, L.; Ponten, F.; Kawaji, H.; Carninci, P.; Forrest, A.R.R.; The FANTOM Consortium; Hayashizaki, Y.; Uhlén, M.; et al. Complementing tissue characterization by integrating transcriptome profiling from the Human Protein Atlas and from the FANTOM5 consortium. Nucleic Acids Res. 2015, 43, 6787–6798. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Kang, B.; Li, C.; Chen, T.; Zhang, Z. GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019, 47, W556–W560. [Google Scholar] [CrossRef]
- Díez-Villanueva, A.; Mallona, I.; Peinado, M.A. Wanderer, an interactive viewer to explore DNA methylation and gene expression data in human cancer. Epigenet. Chromatin 2015, 8, 22. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.E.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef] [PubMed]
- Modhukur, V.; Iljasenko, T.; Metsalu, T.; Lokk, K.; Laisk-Podar, T.; Vilo, J. MethSurv: A web tool to perform multivariable survival analysis using DNA methylation data. Epigenomics 2018, 10, 277–288. [Google Scholar] [CrossRef] [PubMed]
- Andersson, S.; Sundberg, M.; Pristovsek, N.; Ibrahim, A.; Jonsson, P.; Katona, B.; Clausson, C.M.; Zieba, A.; Ramström, M.; Söderberg, O.; et al. Insufficient antibody validation challenges oestrogen receptor beta research. Nat. Commun. 2017, 8, 15840. [Google Scholar] [CrossRef] [PubMed]
- Edwards, D.P. Regulation of signal transduction pathways by estrogen and progesterone. Annu. Rev. Physiol. 2005, 67, 335–376. [Google Scholar] [CrossRef]
- Vivacqua, A.; Bonofiglio, D.; Recchia, A.G.; Musti, A.M.; Picard, D.; Andò, S.; Maggiolini, M. The G protein-coupled receptor GPR30 mediates the proliferative effects induced by 17beta-estradiol and hydroxytamoxifen in endometrial cancer cells. Mol. Endocrinol. 2006, 20, 631–646. [Google Scholar] [CrossRef]
- Périan, S.; Cerutti, C.; Forcet, C.; Tribollet, V.; Vanacker, J.-M. A Cell-Based Method to Detect Agonist and Antagonist Activities of Endocrine-Disrupting Chemicals on GPER. Front. Endocrinol. 2020, 11, 547. [Google Scholar] [CrossRef] [PubMed]
- Saha, S.; Dey, S.; Nath, S. Steroid Hormone Receptors: Links With Cell Cycle Machinery and Breast Cancer Progression. Front. Oncol. 2021, 11, 620214. [Google Scholar] [CrossRef]
- Haynes, B.P.; Schuster, G.; Buus, R.; Alataki, A.; Ginsburg, O.; Quang, L.H.; Han, P.T.; Khoa, P.H.; Van Dinh, N.; Van To, T.; et al. Impact of the menstrual cycle on commercial prognostic gene signatures in oestrogen receptor-positive primary breast cancer. Breast Cancer Res. Treat. 2021, 190, 295–305. [Google Scholar] [CrossRef] [PubMed]
- Ors, A.; Chitsazan, A.D.; Doe, A.R.; Mulqueen, R.M.; Ak, C.; Wen, Y.; Haverlack, S.; Handu, M.; Naldiga, S.; Saldivar, J.C.; et al. Estrogen regulates divergent transcriptional and epigenetic cell states in breast cancer. Nucleic Acids Res. 2022, 50, 11492–11508. [Google Scholar] [CrossRef] [PubMed]
- Irani, S.; Tan, W.; Li, Q.; Toy, W.; Jones, C.; Gadiya, M.; Marra, A.; Katzenellenbogen, J.A.; Carlson, K.E.; Katzenellenbogen, B.S.; et al. Somatic estrogen receptor α mutations that induce dimerization promote receptor activity and breast cancer proliferation. J. Clin. Investig. 2024, 134, e163242. [Google Scholar] [CrossRef] [PubMed]
- Martin, S.G.; Lebot, M.N.; Sukkarn, B.; Ball, G.; Green, A.R.; Rakha, E.A.; Ellis, I.O.; Storr, S.J. Low expression of G protein-coupled oestrogen receptor 1 (GPER) is associated with adverse survival of breast cancer patients. Oncotarget 2018, 9, 25946–25956. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.-L.; Huang, Z.-Y.; Yu, K.; Li, J.; Fu, X.-W.; Deng, S.-L. Estrogen Biosynthesis and Signal Transduction in Ovarian Disease. Front. Endocrinol. 2022, 13, 827032. [Google Scholar] [CrossRef] [PubMed]
- Friese, K.; Kost, B.; Vattai, A.; Marmé, F.; Kuhn, C.; Mahner, S.; Dannecker, C.; Jeschke, U.; Heublein, S. The G protein-coupled estrogen receptor (GPER/GPR30) may serve as a prognostic marker in early-stage cervical cancer. J. Cancer Res. Clin. Oncol. 2018, 144, 13–19. [Google Scholar] [CrossRef]
- Xu, S.; Yu, S.; Dong, D.; Lee, L.T.O. G Protein-Coupled Estrogen Receptor: A Potential Therapeutic Target in Cancer. Front. Endocrinol. 2019, 10, 725. [Google Scholar] [CrossRef]
- Chimento, A.; De Luca, A.; Nocito, M.C.; Avena, P.; La Padula, D.; Zavaglia, L.; Pezzi, V. Role of GPER-Mediated Signaling in Testicular Functions and Tumorigenesis. Cells 2020, 9, 2115. [Google Scholar] [CrossRef]
- Hu, W.-Y.; Shi, G.-B.; Lam, H.-M.; Hu, D.-P.; Ho, S.-M.; Madueke, I.C.; Kajdacsy-Balla, A.; Prins, G.S. Estrogen-initiated transformation of prostate epithelium derived from normal human prostate stem-progenitor cells. Endocrinology 2011, 152, 2150–2163. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Chen, Z.; Jiang, G.; Zhou, Y.; Yang, X.; Huang, H.; Liu, H.; Du, J.; Wang, H. Epigenetic down regulation of G protein-coupled estrogen receptor (GPER) functions as a tumor suppressor in colorectal cancer. Mol. Cancer 2017, 16, 87. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Niu, Y.; Zhang, B.; Sun, Q.; Li, H.; Bai, L.; Su, Z. Different Expression Pattern of G Protein-Coupled Estrogen Receptor GPER1 in Esophageal Squamous Cell Carcinoma and Adenocarcinoma. Int. J. Mol. Sci. 2023, 24, 14055. [Google Scholar] [CrossRef] [PubMed]
- Natale, C.A.; Li, J.; Zhang, J.; Dahal, A.; Dentchev, T.; Stanger, B.Z.; Ridky, T.W. Activation of G protein-coupled estrogen receptor signaling inhibits melanoma and improves response to immune checkpoint blockade. eLife 2018, 7, e31770. [Google Scholar] [CrossRef] [PubMed]
- Edwards, J.R.; O’Donnell, A.H.; Rollins, R.A.; Peckham, H.E.; Lee, C.; Milekic, M.H.; Chanrion, B.; Fu, Y.; Su, T.; Hibshoosh, H.; et al. Chromatin and sequence features that define the fine and gross structure of genomic methylation patterns. Genome Res. 2010, 20, 972–980. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, M.V.C.; Bourc’His, D. The diverse roles of DNA methylation in mammalian development and disease. Nat. Rev. Mol. Cell Biol. 2019, 20, 590–607. [Google Scholar] [CrossRef]
- Gowher, H.; Brick, K.; Camerini-Otero, R.D.; Felsenfeld, G. Vezf1 protein binding sites genome-wide are associated with pausing of elongatingRNApolymeraseII. Proc. Natl. Acad. Sci. USA 2012, 109, 2370–2375. [Google Scholar] [CrossRef] [PubMed]
- Lorincz, M.C.; Dickerson, D.R.; Schmitt, M.; Groudine, M. Intragenic DNAmethylation alters chromatin structure and elongation efficiency in mammalian cells. Nat. Struct. Mol. Biol. 2004, 11, 1068–1075. [Google Scholar] [CrossRef] [PubMed]
- Maunakea, A.K.; Nagarajan, R.P.; Bilenky, M.; Ballinger, T.J.; D’souza, C.; Fouse, S.D.; Johnson, B.E.; Hong, C.; Nielsen, C.; Zhao, Y.; et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature 2010, 466, 253–257. [Google Scholar] [CrossRef]
- Lin, S.; Yi, S.; Qiu, P. Comprehensive analysis of TCGA data reveals correlation between DNA methylation and alternative splicing. BMC Genom. 2022, 23, 758. [Google Scholar] [CrossRef]
- Lewandowski, S.; Kalita, K.; Kaczmarek, L. Estrogen receptor beta. Potential functional significance of a variety of mRNA isoforms. FEBS Lett. 2002, 524, 1–5. [Google Scholar] [CrossRef]
- Métivier, R.; Penot, G.; Carmouche, R.P.; Hübner, M.R.; Reid, G.; Denger, S.; Manu, D.; Brand, H.; Koš, M.; Benes, V.; et al. Transcriptional complexes engaged by apo-estrogen receptor-alpha isoforms have divergent outcomes. EMBO J. 2004, 23, 3653–3666. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.; Dey, P.; Ziegler, Y.; Jiao, X.; Kim, S.H.; Katzenellenbogen, J.A.; Katzenellenbogen, B.S. Contrasting activities of estrogen receptor beta isoforms in triple negative breast cancer. Breast Cancer Res. Treat. 2021, 185, 281–292. [Google Scholar] [CrossRef] [PubMed]
- Skvortsova, K.; Stirzaker, C.; Taberlay, P. The DNA methylation landscape in cancer. Essays Biochem. 2019, 63, 797–811. [Google Scholar]
- Knight, A.K.; Spencer, J.B.; Smith, A.K. DNA methylation as a window into female reproductive aging. Epigenomics 2024, 16, 175–188. [Google Scholar] [CrossRef]
- Zhang, L.-Q.; Liang, Y.-C.; Wang, J.-X.; Zhang, J.; La, T.; Li, Q.-Z. Unlocking the potential: A novel prognostic index signature for acute myeloid leukemia. Comput. Biol. Med. 2024, 173, 108396. [Google Scholar] [CrossRef] [PubMed]
- Kerdivel, G.; Amrouche, F.; Calmejane, M.A.; Carallis, F.; Hamroune, J.; Hantel, C.; Bertherat, J.; Assié, G.; Boeva, V. DNA hypermethylation driven by DNMT1 and DNMT3A favors tumor immune escape contributing to the aggressiveness of adrenocorticalcarcinoma. Clin. Epigenet. 2023, 15, 121. [Google Scholar] [CrossRef]
- Early Breast Cancer Trialists’ Collaborative. Relevance of breast cancer hormone receptors and other factors to the efficacy of adjuvant tamoxifen: Patient-level meta-analysis of randomised trials. Lancet 2011, 378, 771–784. [Google Scholar] [CrossRef]
- Garcia-Martinez, L.; Zhang, Y.; Nakata, Y.; Chan, H.L.; Morey, L. Epigenetic mechanisms in breast cancer therapy and resistance. Nat. Commun. 2021, 12, 1786. [Google Scholar] [CrossRef]
- Sukocheva, O.A.; Lukina, E.; Friedemann, M.; Menschikowski, M.; Hagelgans, A.; Aliev, G. The crucial role of epigenetic regulation in breast cancer anti-estrogen resistance: Current findings and future perspectives. Semin. Cancer Biol. 2022, 82, 35–59. [Google Scholar] [CrossRef]
- Fortin, J.; Chiang, M.F.; Meydan, C.; Foox, J.; Ramachandran, P.; Leca, J.; Lemonnier, F.; Li, W.Y.; Gams, M.S.; Sakamoto, T.; et al. Distinct and opposite effects of leukemogenic Idh and Tet2 mutations in hematopoietic stem and progenitor cells. Proc. Natl. Acad. Sci. USA 2023, 120, e2208176120. [Google Scholar] [CrossRef]
- Sandoval, J.E.; Huang, Y.-H.; Muise, A.; Goodell, M.A.; Reich, N.O. Mutations in the DNMT3A DNA methyltransferase in acute myeloid leukemia patients cause both loss and gain of function and differential regulation by protein partners. J. Biol. Chem. 2019, 294, 4898–4910. [Google Scholar] [CrossRef]
- Zinad, H.S.; Sae-Lee, C.; Ariza-Mateos, M.A.; Adamson, G.; Khazeem, M.M.; Knox, A.; Chung, G.; Mann, J.; Werner, A. Interdependent Transcription of a Natural Sense/Antisense Transcripts Pair (SLC34A1/PFN3). Non-Coding RNA 2022, 8, 19. [Google Scholar] [CrossRef]
- Ali, A.; Khatoon, A.; Shao, C.; Murtaza, B.; Tanveer, Q.; Su, Z. Therapeutic potential of natural antisense transcripts and various mechanisms involved for clinical applications and disease prevention. RNA Biol. 2024, 21, 1–18. [Google Scholar] [CrossRef]



| Tumor (Number of Patients/Samples) | ESR1 | ESR2 | GPER1 |
|---|---|---|---|
| Adrenocortical carcinoma (88) | Pearson: −0.47 (p = 2.061 × 10−5) | Pearson: −0.49 (p = 7.873 × 10−6) | Pearson: −0.74 (p = 5.08 × 10−14) |
| Bladder urothelial carcinoma (127) | Pearson: −0.14 (p = 0.126) | Pearson: −0.18 (p = 0.0436) | Pearson: −0.36 (p = 3.259 × 10−5) |
| Breast invasive carcinoma (963) | Pearson: −0.74 (p = 2.52 × 10−116) | Pearson: −0.01 (p = 0.846) | Pearson: −0.36 p = 8.62 × 10−22) |
| Cervical squamous cell carcinoma and endocervical adenocarcinoma (191) | Pearson: −0.54 (p = 5.40 × 10−16) | Pearson: −0.05 (p = 0.505) | Pearson: −0.48 (p = 1.56 × 10−12) |
| Cholangiocarcinoma (35) | Pearson: −0.33 (p = 0.0551) | Pearson: −0.28 (p = 0.101) | Pearson: −0.67 (p = 9.217 × 10−6) |
| Esophageal carcinoma (184) | Pearson: −0.45 (p = 1.09 × 10−10) | Pearson: −0.09 (p = 0.213) | Pearson: −0.41 (p = 8.76 × 10−9) |
| Head and neck squamous cell carcinoma (504) | Pearson: −0.18 (p = 8.349 × 10−5) | Pearson: −0.23 (p = 3.90 × 10−7) | Pearson: −0.49 (p = 8.74 × 10−31) |
| Kidney chromophobe (66) | Pearson: −0.24 (p = 0.0557) | Pearson: −0.03 (p = 0.836) | Pearson: −0.47 (p = 7.333 × 10−5) |
| Kidney renal clear cell carcinoma (448) | Pearson: −0.41 (p = 3.48 × 10−12) | Pearson: −0.26 (p =2.622 × 10−5) | Pearson: −0.43 (p = 4.82 × 10−13) |
| Kidney renal papillary cell carcinoma (280) | Pearson: −0.56 (p = 4.33 × 10−23) | Pearson: −0.18 (p = 3.705 × 10−3) | Pearson: −0.63 (p = 1.14 × 10−30) |
| Lower-grade glioma (283) | Pearson: −0.51 (p = 7.10 × 10−20) | Pearson: −0.04 (p = 0.489) | Pearson: −0.26 (p = 1.055 × 10−5) |
| Liver hepatocellular carcinoma (366) | Pearson: 0.04 (p = 0.492) | Pearson: −0.34 (p = 2.49 × 10−11) | Pearson: −0.62 (p = 4.99 × 10−39) |
| Lung adenocarcinoma (230) | Pearson: −0.40 (p = 1.33 × 10−8) | Pearson: −0.13 (p = 0.0729) | Pearson: −0.38 (p = 8.56 × 10−8) |
| Lung squamous cell carcinoma (178) | Pearson: −0.11 (p = 0.344) | Pearson: −0.25 (p = 0.0309) | Pearson: −0.18 (p = 0.128) |
| Pancreatic adenocarcinoma (149) | Pearson: −0.31 (p = 1.339 × 10−4) | Pearson: −0.14 (p = 0.0982) | Pearson: −0.47 (p = 1.00 × 10−9) |
| Prostate adenocarcinoma (492) | Pearson: −0.36 (p = 2.28 × 10−16) | Pearson: −0.18 (p = 6.165 × 10−5) | Pearson: −0.57 (p = 1.00 × 10−43) |
| Thyroid carcinoma (399) | Pearson: −0.54 (p = 3.82 × 10−31) | Pearson: −0.03 (p = 0.548) | Pearson: −0.32 (p = 1.21 × 10−10) |
| Thymoma (123) | Pearson: −0.37 (p = 3.940 × 10−5) | Pearson: −0.49 (p = 1.86 × 10−8) | Pearson: −0.31 (p = 7.031 × 10−4) |
| Uterine carcinosarcoma (56) | Pearson: −0.18 (p = 0.185) | Pearson: −0.04 (p = 0.792) | Pearson: −0.72 (p = 6.00 × 10−10) |
| Name. | Cancer * | HR | CI | p-Value | To Ref. Gene # | Island # |
|---|---|---|---|---|---|---|
| ESR1 | ||||||
| cg25490334 | UVM | 0.077 | (0.01; 0.578) | 0.012625 | Body | Open_Sea |
| cg08907436 | KIRC | 0.159 | (0.07; 0.363) | 1.28 × 10−5 | 5′UTR; TSS1500 | N_Shelf |
| cg01715172 | UVM | 0.186 | (0.043; 0.809) | 0.024897 | TSS1500; 5′UTR | N_Shore |
| cg20893956 | UVM | 0.231 | (0.101; 0.529) | 0.000518 | 5′UTR; TSS200 | N_Shelf |
| cg21265702 | UVM | 0.232 | (0.086; 0.626) | 0.003911 | Body | Open_Sea |
| cg19411146 | KICH | 6.807 | (1.696; 27.319) | 0.006824 | 5′UTR; 1stExon | N_Shore |
| cg18745416 | UVM | 10.649 | (2.451; 46.269) | 0.001599 | TSS1500 | Open_Sea |
| cg20253551 | KICH | 11.207 | (2.325; 54.026) | 0.002603 | Body; 1stExon | CG Island |
| cg25338972 | UVM | 13.029 | (2.995; 56.676) | 0.000621 | TSS1500 | Open_Sea |
| cg06611115 | UVM | 20.689 | (2.734; 156.579) | 0.003348 | TSS1500 | Open_Sea |
| ESR2 | ||||||
| cg22778790 | UVM | 0.072 | (0.01; 0.541) | 0.010473 | TSS1500; 5′UTR | S_Shore |
| cg23494957 | UVM | 0.102 | (0.014; 0.759) | 0.025805 | TSS1500; 5′UTR | S_Shore |
| cg01845091 | UVM | 0.209 | (0.049; 0.895) | 0.034904 | TSS1500; 5′UTR | S_Shore |
| cg16792632 | KICH | 0.237 | (0.063; 0.887) | 0.032475 | TSS200; 5′UTR | CG Island |
| cg20019356 | UVM | 0.24 | (0.105; 0.547) | 0.000683 | 5′UTR | S_Shelf |
| cg23284245 | UCEC | 2.782 | (1.576; 4.913) | 0.000419 | 5′UTR | N_Shelf |
| cg01845091 | ACC | 2.786 | (1.301; 5.965) | 0.008339 | TSS1500; 5′UTR | S_Shore |
| cg19583967 | ACC | 2.863 | (1.293; 6.343) | 0.009529 | TSS1500 | S_Shore |
| cg11059483 | KICH | 3.962 | (1.061; 14.802) | 0.0406 | 1stExon; 5′UTR | Island |
| cg19583967 | UVM | 4.51 | (1.055; 19.285) | 0.042191 | TSS1500 | S_Shore |
| GPER1 | ||||||
| cg06449934 | UVM | 0.083 | (0.011; 0.653) | 0.018014 | 5′UTR; Body | N_Shore |
| cg16127845 | LGG | 0.149 | (0.102; 0.216) | 0 | TSS200; Body | N_Shore |
| cg18378883 | LGG | 0.153 | (0.105; 0.223) | 0 | TSS200; Body | N_Shore |
| cg17102910 | LGG | 0.197 | (0.136; 0.284) | 0 | TSS200; Body | N_Shore |
| cg01504489 | UVM | 0.227 | (0.084; 0.612) | 0.003393 | Body | CG Island |
| cg07904865 | UVM | 5.288 | (1.232; 22.694) | 0.025032 | 5′UTR; Body | S_Shore |
| cg03595161 | UVM | 5.362 | (1.243; 23.119) | 0.024311 | 5′UTR; Body | S_Shore |
| cg19723715 | ACC | 6.093 | (1.445; 25.682) | 0.01382 | TSS1500; Body | N_Shore |
| cg15730481 | UVM | 10.298 | (1.383; 76.672) | 0.022812 | TSS200; Body | N_Shore |
| cg17102910 | ACC | 13.854 | (1.878; 102.186) | 0.009931 | TSS200; Body | N_Shore |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rong, J.; Xie, X.; Niu, Y.; Su, Z. Correlation between the RNA Expression and the DNA Methylation of Estrogen Receptor Genes in Normal and Malignant Human Tissues. Curr. Issues Mol. Biol. 2024, 46, 3610-3625. https://doi.org/10.3390/cimb46040226
Rong J, Xie X, Niu Y, Su Z. Correlation between the RNA Expression and the DNA Methylation of Estrogen Receptor Genes in Normal and Malignant Human Tissues. Current Issues in Molecular Biology. 2024; 46(4):3610-3625. https://doi.org/10.3390/cimb46040226
Chicago/Turabian StyleRong, Ju, Xiaojun Xie, Yongdong Niu, and Zhongjing Su. 2024. "Correlation between the RNA Expression and the DNA Methylation of Estrogen Receptor Genes in Normal and Malignant Human Tissues" Current Issues in Molecular Biology 46, no. 4: 3610-3625. https://doi.org/10.3390/cimb46040226





