Effects of Lignin-Diverted Reductant with Polyphenol Oxidases on Cellulose Degradation by Wild and Mutant Types of Lytic Polysaccharide Monooxygenase
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Expression and Purification of Lytic Polysaccharide Monooxygenases (LMPOs)
2.3. Assessment of Lytic Polysaccharide Monooxygenase (LMPO) Activity
2.4. Simultaneous Reactions of Lytic Polysaccharide Monooxygenase (LMPO) and Polyphenol Oxidase
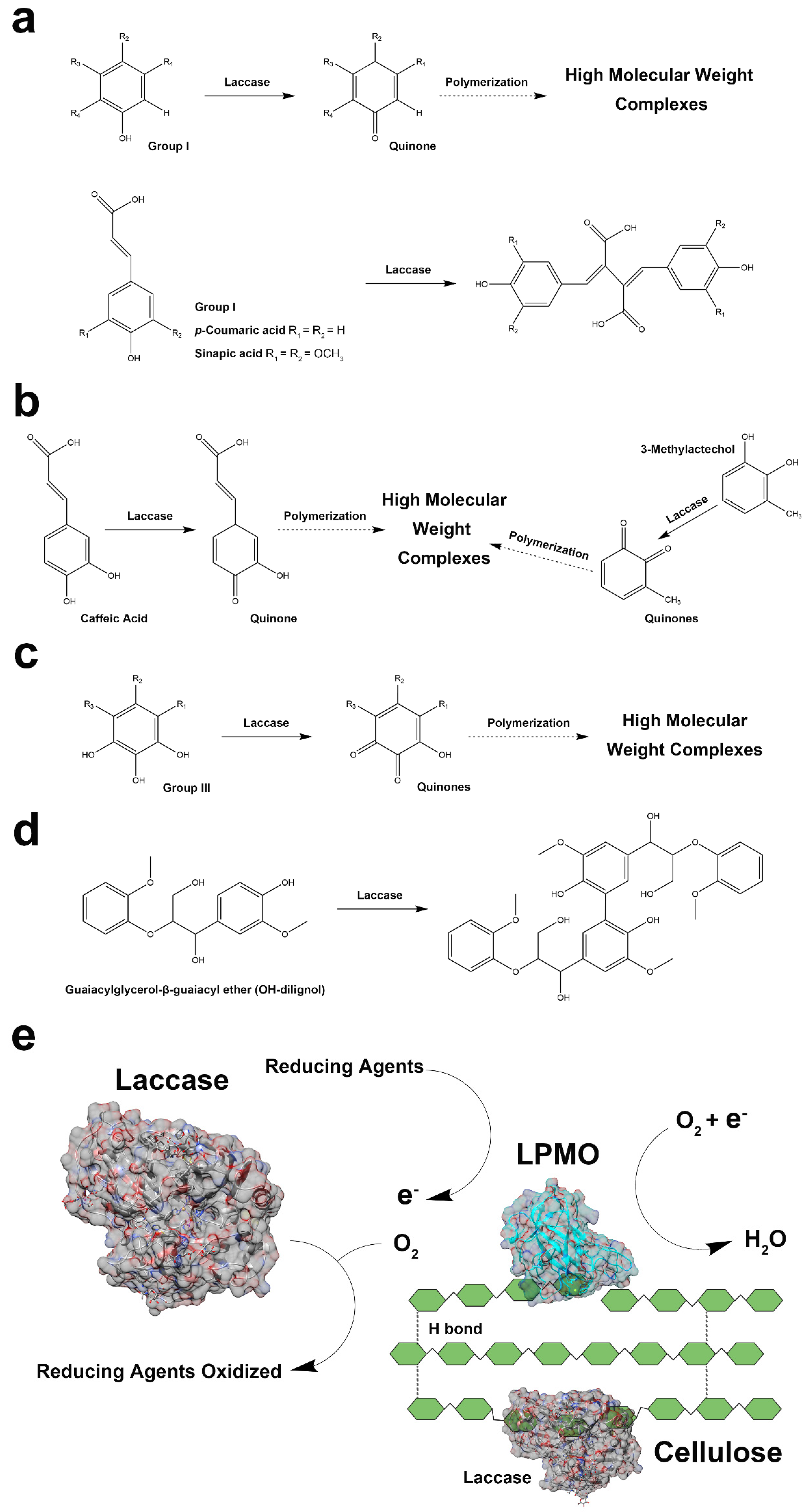
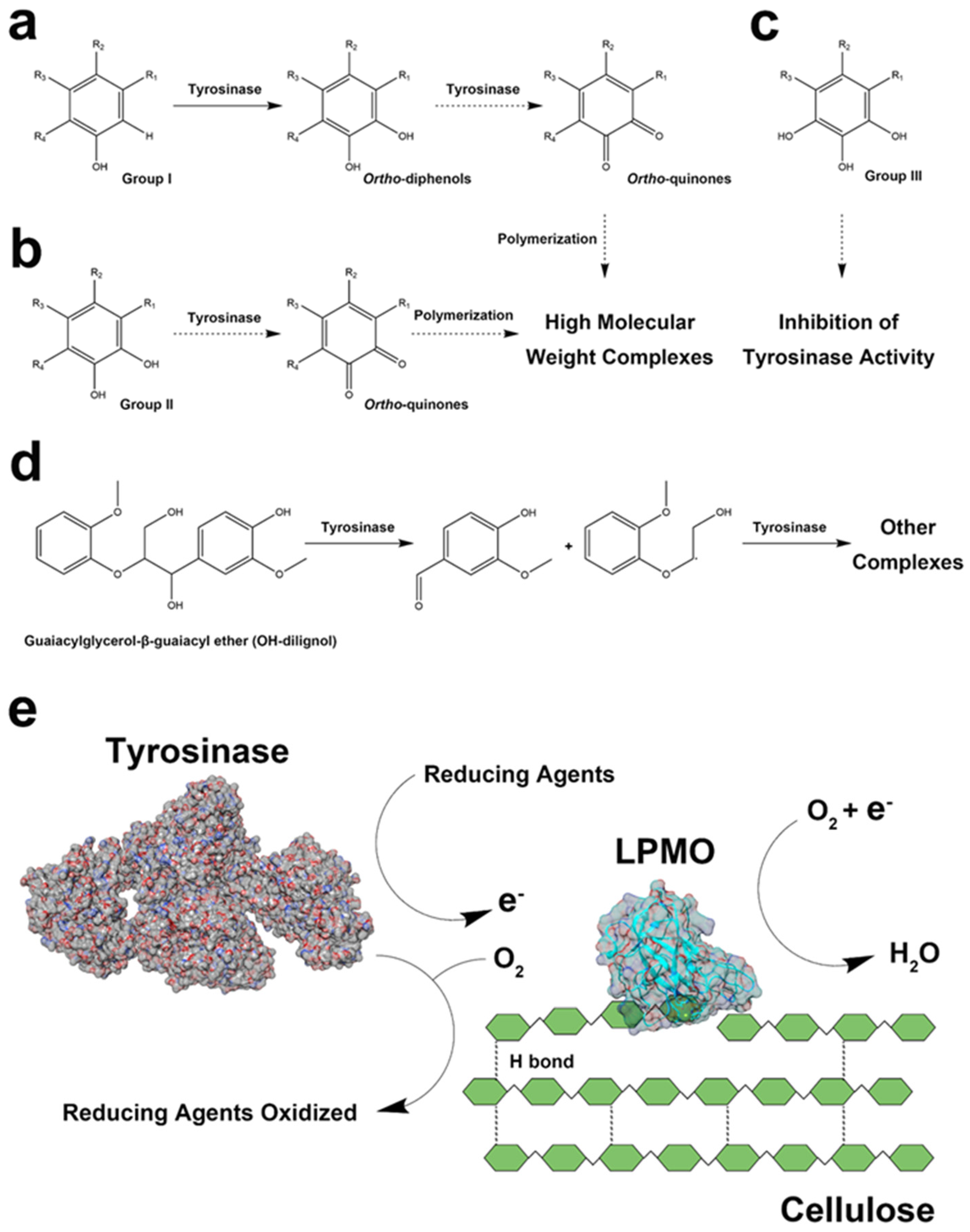
2.5. Oxidation Products of Laccase and Tyrosinase
2.6. Protein Structures and Electrostatic Plots of Lytic Polysaccharide Monooxygenases (LMPOs)
2.7. Optimal pH and Temperature Conditions of Laccase and Tyrosinase
2.8. Kinetic Parameters of Laccase and Tyrosinase
2.9. Statistical Analysis
3. Results and Discussion
3.1. Simultaneous Reactions of Lytic Polysaccharide Monooxygenase (LMPO) and Polyphenol Oxidase
3.2. Evaluation of Lytic Polysaccharide Monooxygenase (LMPO) Activities
3.2.1. Monophenols Group Reducing Agents
3.2.2. Other Reducing Agents
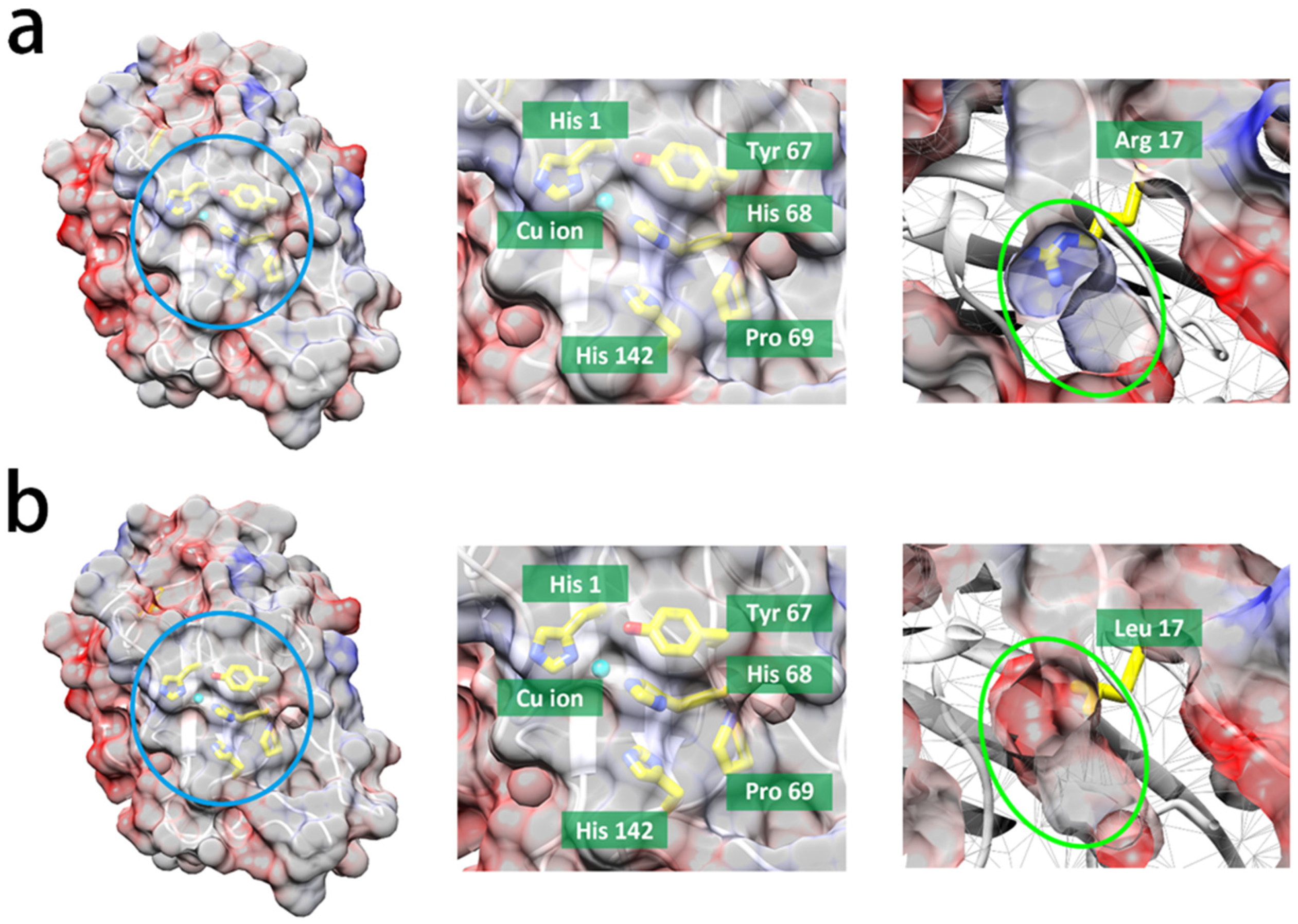
3.3. Insights into Protein Structure and Reducing Agent Specificity for Lytic Polysaccharide Monooxygenase (LMPO) Reactions
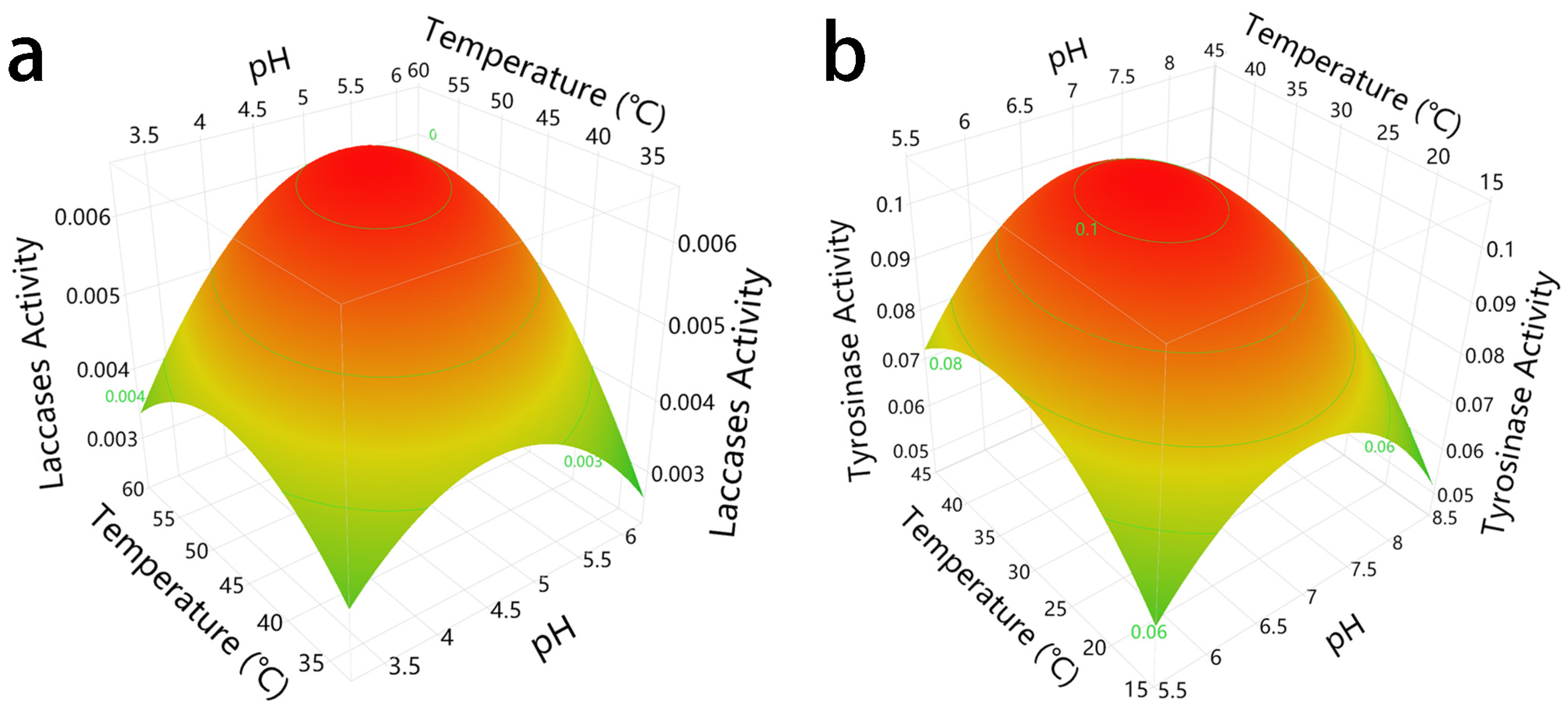
3.4. Optimal Reaction Conditions of Laccase and Tyrosinase
3.5. Kinetic Parameters of Laccase and Tyrosinase
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Munk, L.; Andersen, M.L.; Meyer, A.S. Direct rate assessment of laccase catalysed radical formation in lignin by electron paramagnetic resonance spectroscopy. Enzym. Microb. Technol. 2017, 106, 88–96. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Lee, H.W.; Lee, S.M.; Jae, J.; Park, Y.K. Overview of the recent advances in lignocellulose liquefaction for producing biofuels, bio-based materials and chemicals. Bioresour. Technol. 2019, 279, 373–384. [Google Scholar] [CrossRef] [PubMed]
- Cheng, K. Industrial scale lignin recovery from pulping liquors. In Natural Polyphenols from Wood; Elsevier: Amsterdam, The Netherlands, 2021; pp. 123–145. [Google Scholar]
- Frommhagen, M.; Mutte, S.K.; Westphal, A.H.; Koetsier, M.J.; Hinz, S.W.A.; Visser, J.; Vincken, J.P.; Weijers, D.; Van Berkel, W.J.H.; Gruppen, H.; et al. Boosting LPMO-driven lignocellulose degradation by polyphenol oxidase-activated lignin building blocks. Biotechnol. Biofuels 2017, 10, 121. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Jiang, X.; Shen, X.; Hu, J.; Tang, W.; Wu, X.; Ragauskas, A.; Jameel, H.; Meng, X.; Yong, Q. Lignin-enzyme interaction: A roadblock for efficient enzymatic hydrolysis of lignocellulosics. Renew. Sustain. Energy Rev. 2022, 154, 111822. [Google Scholar] [CrossRef]
- Ren, S.; Li, C.; Jiao, X.; Jia, S.; Jiang, Y.; Bilal, M.; Cui, J. Recent progress in multienzymes co-immobilization and multienzyme system applications. Chem. Eng. J. 2019, 373, 1254–1278. [Google Scholar] [CrossRef]
- Kim, S.; Ståhlberg, J.; Sandgren, M.; Paton, R.S.; Beckham, G.T. Quantum mechanical calculations suggest that lytic polysaccharide monooxygenases use a copper-oxyl, oxygen-rebound mechanism. Proc. Natl. Acad. Sci. USA 2014, 111, 149–154. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Zhang, J.; Ma, F.; Chen, Q.; Xiao, Q.; Zhang, X.; Xie, S.; Yu, H. Lytic polysaccharide monooxygenases promote oxidative cleavage of lignin and lignin–carbohydrate complexes during fungal degradation of lignocellulose. Environ. Microbiol. 2021, 23, 4547–4560. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; An, Y.; Chai, C.; Sang, J.; Jiang, L.; Lu, F.; Dai, Y.; Liu, F. Construction of the R17L mutant of MtC1LPMO for improved lignocellulosic biomass conversion by rational point mutation and investigation of the mechanism by molecular dynamics simulations. Bioresour. Technol. 2020, 317, 124024. [Google Scholar] [CrossRef] [PubMed]
- Levasseur, A.; Drula, E.; Lombard, V.; Coutinho, P.M.; Henrissat, B. Expansion of the enzymatic repertoire of the CAZy database to integrate auxiliary redox enzymes. Biotechnol. Biofuels 2013, 6, 41. [Google Scholar] [CrossRef]
- Wu, S.; Tian, J.; Xie, N.; Adnan, M.; Wang, J.; Liu, G. A sensitive, accurate, and high-throughput gluco-oligosaccharide oxidase-based HRP colorimetric method for assaying lytic polysaccharide monooxygenase activity. Biotechnol. Biofuels Bioprod. 2022, 15, 15. [Google Scholar] [CrossRef]
- Hemsworth, G.R.; Johnston, E.M.; Davies, G.J.; Walton, P.H. Lytic Polysaccharide Monooxygenases in Biomass Conversion. Trends Biotechnol. 2015, 33, 747–761. [Google Scholar] [CrossRef] [PubMed]
- Sabbadin, F.; Hemsworth, G.R.; Ciano, L.; Henrissat, B.; Dupree, P.; Tryfona, T.; Marques, R.D.S.; Sweeney, S.T.; Besser, K.; Elias, L.; et al. An ancient family of lytic polysaccharide monooxygenases with roles in arthropod development and biomass digestion. Nat. Commun. 2018, 9, 756. [Google Scholar] [CrossRef] [PubMed]
- Janusz, G.; Pawlik, A.; Świderska-Burek, U.; Polak, J.; Sulej, J.; Jarosz-Wilkołazka, A.; Paszczyński, A. Laccase Properties, Physiological Functions, and Evolution. Int. J. Mol. Sci. 2020, 21, 966. [Google Scholar] [CrossRef] [PubMed]
- Perna, V.; Meyer, A.S.; Holck, J.; Eltis, L.D.; Eijsink, V.G.H.; Wittrup Agger, J. Laccase-Catalyzed Oxidation of Lignin Induces Production of H 2 O 2. ACS Sustain. Chem. Eng. 2020, 8, 831–841. [Google Scholar] [CrossRef]
- Long, L.; Hu, Y.; Sun, F.; Gao, W.; Hao, Z.; Yin, H. Advances in lytic polysaccharide monooxygenases with the cellulose-degrading auxiliary activity family 9 to facilitate cellulose degradation for biorefinery. Int. J. Biol. Macromol. 2022, 219, 68–83. [Google Scholar] [CrossRef] [PubMed]
- Westereng, B.; Cannella, D.; Wittrup Agger, J.; Jørgensen, H.; Larsen Andersen, M.; Eijsink, V.G.H.; Felby, C. Enzymatic cellulose oxidation is linked to lignin by long-range electron transfer. Sci. Rep. 2015, 5, 18561. [Google Scholar] [CrossRef] [PubMed]
- Brenelli, L.; Squina, F.M.; Felby, C.; Cannella, D. Laccase-derived lignin compounds boost cellulose oxidative enzymes AA9. Biotechnol. Biofuels 2018, 11, 10. [Google Scholar] [CrossRef] [PubMed]
- Gerbin, E.; Frapart, Y.-M.; Marcuello, C.; Cottyn, B.; Foulon, L.; Pernes, M.; Crônier, D.; Molinari, M.; Chabbert, B.; Ducrot, P.-H.; et al. Dual Antioxidant Properties and Organic Radical Stabilization in Cellulose Nanocomposite Films Functionalized by In Situ Polymerization of Coniferyl Alcohol. Biomacromolecules 2020, 21, 3163–3175. [Google Scholar] [CrossRef] [PubMed]
- Min, K.; Yum, T.; Kim, J.; Woo, H.M.; Kim, Y.; Sang, B.-I.; Yoo, Y.J.; Kim, Y.H.; Um, Y. Perspectives for biocatalytic lignin utilization: Cleaving 4-O-5 and Cα–Cβ bonds in dimeric lignin model compounds catalyzed by a promiscuous activity of tyrosinase. Biotechnol. Biofuels 2017, 10, 212. [Google Scholar] [CrossRef]
- Muraleedharan, M.N.; Zouraris, D.; Karantonis, A.; Topakas, E.; Sandgren, M.; Rova, U.; Christakopoulos, P.; Karnaouri, A. Effect of lignin fractions isolated from different biomass sources on cellulose oxidation by fungal lytic polysaccharide monooxygenases. Biotechnol. Biofuels 2018, 11, 296. [Google Scholar] [CrossRef]
- Guo, X.; Sang, J.; Chai, C.; An, Y.; Wei, Z.; Zhang, H.; Ma, L.; Dai, Y.; Lu, F.; Liu, F. A lytic polysaccharide monooxygenase from Myceliophthora thermophila C1 and its characterization in cleavage of glycosidic chain of cellulose. Biochem. Eng. J. 2020, 162, 107712. [Google Scholar] [CrossRef]
- Miller, G.L. Use of Dinitrosalicylic Acid Reagent for Determination of Reducing Sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [PubMed]
- Tan, T.-C.; Kracher, D.; Gandini, R.; Sygmund, C.; Kittl, R.; Haltrich, D.; Hällberg, B.M.; Ludwig, R.; Divne, C. Structural basis for cellobiose dehydrogenase action during oxidative cellulose degradation. Nat. Commun. 2015, 6, 7542. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera? A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- d’Acunzo, F.; Galli, C.; Gentili, P.; Sergi, F. Mechanistic and steric issues in the oxidation of phenolic and non-phenolic compounds by laccase or laccase-mediator systems. The case of bifunctional substrates. New J. Chem. 2006, 30, 583. [Google Scholar] [CrossRef]
- Frommhagen, M.; Koetsier, M.J.; Westphal, A.H.; Visser, J.; Hinz, S.W.A.; Vincken, J.-P.; van Berkel, W.J.H.; Kabel, M.A.; Gruppen, H. Lytic polysaccharide monooxygenases from Myceliophthora thermophila C1 differ in substrate preference and reducing agent specificity. Biotechnol. Biofuels 2016, 9, 186. [Google Scholar] [CrossRef]
- de Oliveira Gorgulho Silva, C.; Vuillemin, M.; Kabel, M.A.; van Berkel, W.J.H.; Meyer, A.S.; Agger, J.W. Polyphenol Oxidase Products Are Priming Agents for LPMO Peroxygenase Activity. ChemSusChem 2023, 16, e202300559. [Google Scholar] [CrossRef] [PubMed]
- Sun, P.; Huang, Z.; Banerjee, S.; Kadowaki, M.A.S.; Veersma, R.J.; Magri, S.; Hilgers, R.; Muderspach, S.J.; Laurent, C.V.F.P.; Ludwig, R.; et al. AA16 Oxidoreductases Boost Cellulose-Active AA9 Lytic Polysaccharide Monooxygenases from Myceliophthora thermophila. ACS Catal. 2023, 13, 4454–4467. [Google Scholar] [CrossRef]
- Kracher, D.; Scheiblbrandner, S.; Felice, A.K.G.; Breslmayr, E.; Preims, M.; Ludwicka, K.; Haltrich, D.; Eijsink, V.G.H.; Ludwig, R. Extracellular electron transfer systems fuel cellulose oxidative degradation. Science 2016, 352, 1098–1101. [Google Scholar] [CrossRef]
- Ingold, C.K. Principles of an Electronic Theory of Organic Reactions. Chem. Rev. 1934, 15, 225–274. [Google Scholar] [CrossRef]
- Steenken, S.; Neta, P. One-electron redox potentials of phenols. Hydroxy- and aminophenols and related compounds of biological interest. J. Phys. Chem. 1982, 86, 3661–3667. [Google Scholar] [CrossRef]
- Hansch, C.; Leo, A.; Taft, R.W. A survey of Hammett substituent constants and resonance and field parameters. Chem. Rev. 1991, 91, 165–195. [Google Scholar] [CrossRef]
- Braunschmid, V.; Binder, K.; Fuerst, S.; Subagia, R.; Danner, C.; Weber, H.; Schwaiger, N.; Nyanhongo, G.S.; Ribitsch, D.; Guebitz, G.M. Comparison of a fungal and a bacterial laccase for lignosulfonate polymerization. Process Biochem. 2021, 109, 207–213. [Google Scholar] [CrossRef]
- Boonchuay, P.; Techapun, C.; Seesuriyachan, P.; Chaiyaso, T. Production of xylooligosaccharides from corncob using a crude thermostable endo-xylanase from Streptomyces thermovulgaris TISTR1948 and prebiotic properties. Food Sci. Biotechnol. 2014, 23, 1515–1523. [Google Scholar] [CrossRef]
- Hahn, V.; Mikolasch, A.; Kuhlisch, C.; Schauer, F. Laccase-mediated multi-step homo- and heteromolecular reactions of ortho-dihydroxylated aromatic compounds and mono- or diaminated substances resulting in C-C, C-O and C-N bonds. J. Mol. Catal. B Enzym. 2015, 122, 56–63. [Google Scholar] [CrossRef]
- Tarrago, L.; Modolo, C.; Yemloul, M.; Robert, V.; Rousselot-Pailley, P.; Tron, T. Controlling the polymerization of coniferyl alcohol with cyclodextrins. New J. Chem. 2018, 42, 11770–11775. [Google Scholar] [CrossRef]
- Nemadziva, B.; Le Roes-Hill, M.; Koorbanally, N.; Kudanga, T. Small laccase-catalyzed synthesis of a caffeic acid dimer with high antioxidant capacity. Process Biochem. 2018, 69, 99–105. [Google Scholar] [CrossRef]
- Perna, V.; Agger, J.W.; Holck, J.; Meyer, A.S. Multiple Reaction Monitoring for quantitative laccase kinetics by LC-MS. Sci. Rep. 2018, 8, 8114. [Google Scholar] [CrossRef]
- Cardullo, N.; Muccilli, V.; Tringali, C. Laccase-mediated synthesis of bioactive natural products and their analogues. RSC Chem. Biol. 2022, 3, 614–647. [Google Scholar] [CrossRef]
- Wang, H.; You, S.; Wang, W.; Zeng, Y.; Su, R.; Qi, W.; Wang, K.; He, Z. Laccase-catalyzed soy protein and gallic acid complexation: Effects on conformational structures and antioxidant activity. Food Chem. 2022, 375, 131865. [Google Scholar] [CrossRef] [PubMed]
- Slagman, S.; Escorihuela, J.; Zuilhof, H.; Franssen, M.C.R. Characterization of the laccase-mediated oligomerization of 4-hydroxybenzoic acid. RSC Adv. 2016, 6, 99367–99375. [Google Scholar] [CrossRef]
- Kuusk, S.; Eijsink, V.G.H.; Väljamäe, P. The “life-span” of lytic polysaccharide monooxygenases (LPMOs) correlates to the number of turnovers in the reductant peroxidase reaction. J. Biol. Chem. 2023, 299, 105094. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.; Fan, L.; Duan, Z. Five individual polyphenols as tyrosinase inhibitors: Inhibitory activity, synergistic effect, action mechanism, and molecular docking. Food Chem. 2019, 297, 124910. [Google Scholar] [CrossRef] [PubMed]
- Kalia, S.; Thakur, K.; Kumar, A.; Celli, A. Laccase-assisted surface functionalization of lignocellulosics. J. Mol. Catal. B Enzym. 2014, 102, 48–58. [Google Scholar] [CrossRef]
- Moilanen, U.; Kellock, M.; Galkin, S.; Viikari, L. The laccase-catalyzed modification of lignin for enzymatic hydrolysis. Enzym. Microb. Technol. 2011, 49, 492–498. [Google Scholar] [CrossRef] [PubMed]
- Munk, L.; Sitarz, A.K.; Kalyani, D.C.; Mikkelsen, J.D.; Meyer, A.S. Can laccases catalyze bond cleavage in lignin? Biotechnol. Adv. 2015, 33, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Moilanen, U.; Kellock, M.; Várnai, A.; Andberg, M.; Viikari, L. Mechanisms of laccase-mediator treatments improving the enzymatic hydrolysis of pre-treated spruce. Biotechnol. Biofuels 2014, 7, 177. [Google Scholar] [CrossRef] [PubMed]
- Oliva-Taravilla, A.; Tomás-Pejó, E.; Demuez, M.; González-Fernández, C.; Ballesteros, M. Phenols and lignin: Key players in reducing enzymatic hydrolysis yields of steam-pretreated biomass in presence of laccase. J. Biotechnol. 2016, 218, 94–101. [Google Scholar] [CrossRef]
- Jin, X.; Yu, X.; Zhu, G.; Zheng, Z.; Feng, F.; Zhang, Z. Conditions Optimizing and Application of Laccase-mediator System (LMS) for the Laccase-catalyzed Pesticide Degradation. Sci. Rep. 2016, 6, 35787. [Google Scholar] [CrossRef]
- Zaidi, K.U.; Ali, A.S.; Ali, S.A. Purification and Characterization of Melanogenic Enzyme Tyrosinase from Button Mushroom. Enzym. Res. 2014, 2014, 120739. [Google Scholar] [CrossRef] [PubMed]
- Zeuner, B.; Thomsen, T.B.; Stringer, M.A.; Krogh, K.B.R.M.; Meyer, A.S.; Holck, J. Comparative Characterization of Aspergillus Pectin Lyases by Discriminative Substrate Degradation Profiling. Front. Bioeng. Biotechnol. 2020, 8, 873. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.-M.; Patel, A.K.; Singhania, R.R.; Tsai, C.-H.; Chen, S.-Y.; Chen, C.-W.; Dong, C. Di Heterologous expression of bacterial CotA-laccase, characterization and its application for biodegradation of malachite green. Bioresour. Technol. 2021, 340, 125708. [Google Scholar] [CrossRef] [PubMed]
- Ramírez-Cavazos, L.I.; Junghanns, C.; Ornelas-Soto, N.; Cárdenas-Chávez, D.L.; Hernández-Luna, C.; Demarche, P.; Enaud, E.; García-Morales, R.; Agathos, S.N.; Parra, R. Purification and characterization of two thermostable laccases from Pycnoporus sanguineus and potential role in degradation of endocrine disrupting chemicals. J. Mol. Catal. B Enzym. 2014, 108, 32–42. [Google Scholar] [CrossRef]
- Zynek, K.; Bryjak, J.; Polakovič, M. Effect of separation on thermal stability of tyrosinase from Agaricus bisporus. J. Mol. Catal. B Enzym. 2010, 66, 172–176. [Google Scholar] [CrossRef]

| Group | Reducing Agents | Activity of R17L in Sodium Ascorbate Set to 100% | |||||
|---|---|---|---|---|---|---|---|
| WT | WT + Laccase | WT + Tyrosinase | R17L | R17L + Laccase | R17L + Tyrosinase | ||
| 1a | 4-Hydroxybenzoic acid | 73.6 ± 4.4 c | 16.6 ± 12.6 e | 69.5 ± 7.9 c | 70.3 ± 5.2 c | 42.4 ± 6.1 d | 95.9 ± 3.7 ab |
| 1a | p-Coumaric acid | 51.6 ± 2.2 c | 62.8 ± 4.4 bc | 79.1 ± 4.2 b | 45.5 ± 1.7 c | 74.6 ± 9.4 b | 102.7 ± 19.1 a |
| 1b | Coniferyl alcohol | 77.6 ± 0.7 bc | 19.6 ± 1.8 d | 90.1 ± 1.8 ab | 85.2 ± 10.0 b | 12.1 ± 5.5 d | 63.9 ± 3.5 c |
| 1b | Guaiacol | 60.7 ± 1.0 d | 69.4 ± 2.7 c | 71.1 ± 3.3 c | 61.3 ± 6.1 d | 65.4 ± 5.0 cd | 57.9 ± 1.1 d |
| 1b | Vanillic acid | 63.5 ± 0.6 c | 8.20 ± 1.2 f | 52.0 ± 3.6 d | 64.3 ± 0.8 c | 7.10 ± 0.3 f | 43.2 ± 7.6 e |
| 1b | 3-Hydroxy-4-methoxycinnamic acid | 66.4 ± 1.8 c | 17.3 ± 1.0 e | 71.6 ± 6.5 c | 70.1 ± 4.9 c | 28.4 ± 6.2 d | 69.3 ± 1.8 c |
| 1b | 4-Hydroxy-3-methoxycinnamic acid | 64.7 ± 8.5 c | 21.3 ± 5.7 d | 60.1 ± 0.5 c | 60.3 ± 2.3 c | 29.5 ± 5.0 d | 69.4 ± 5.3 c |
| 1b | 4-Hydroxy-3-methoxyphenylacetone | 60.1 ± 0.8 e | 10.9 ± 0.1 g | 88.4 ± 4.2 b | 64.3 ± 3.5 e | 26.1 ± 1.3 f | 71.0 ± 0.4 d |
| 1b | 4-Hydroxy-3-methoxyphenylacetic acid | 76.0 ± 1.8 bc | 31.8 ± 6.0 e | 73.7 ± 3.7 bc | 66.6 ± 6.7 c | 43.2 ± 3.3 d | 70.2 ± 6.2 c |
| 1c | Sinapic acid | 69.9 ± 2.7 e | 86.3 ± 5.3 bc | 79.4 ± 7.0 cde | 71.6 ± 3.3 de | 80.8 ± 1.0 bcd | 90.9 ± 5.9 ab |
| 2 | 3-Methylcatechol | 96.8 ± 6.2 bc | 110.9 ± 0.7 a | 96.0 ± 2.0 bc | 100.7 ± 5.0 b | 103.6 ± 4.2 ab | 90.9 ± 4.3 cd |
| 2 | Caffeic acid | 83.7 ± 3.4 cd | 31.8 ± 1.4 e | 69.7 ± 4.7 d | 98.7 ± 15.1 ab | 26.6 ± 1.2 e | 84.8 ± 6.5 bc |
| 3 | Gallic acid | 111.3 ± 1.0 b | 77.1 ± 13.7 c | 99.1 ± 5.4 bc | 113.0 ± 1.6 b | 171.3 ± 30.0 a | 152.3 ± 5.9 a |
| other | Guaiacylglycerol-β-guaiacyl ether | 59.0 ± 4.4 bc | 44.9 ± 3.7 c | 60.4 ± 4.4 b | 63.4 ± 9.9 b | 53.4 ± 2.7 bc | 60.4 ± 4.4 b |
| Name | Vmax (mM/min) | Km (mM) | kcat (/min) | kcat/Km (mM−1 min−1) | Temperature (°C) | Kd (min−1) | t½ (min) |
|---|---|---|---|---|---|---|---|
| Laccase | 0.003827 | 0.1004 | 14.12 | 140.6 | 65 | 0.01300 | 53.32 |
| 70 | 0.05032 | 13.77 | |||||
| 75 | 0.06771 | 10.24 | |||||
| 80 | 0.08381 | 8.27 | |||||
| 85 | 0.2227 | 3.11 | |||||
| Tyrosinase | 0.000007632 | 0.2440 | 0.04270 | 0.1750 | 50 | 0.01115 | 62.17 |
| 55 | 0.01624 | 42.68 | |||||
| 60 | 0.04393 | 15.78 | |||||
| 65 | 0.05774 | 12.00 | |||||
| 70 | 0.1732 | 4.00 | |||||
| 75 | 0.2603 | 2.66 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, K.; Wang, Y.; Guo, X.; Wang, B. Effects of Lignin-Diverted Reductant with Polyphenol Oxidases on Cellulose Degradation by Wild and Mutant Types of Lytic Polysaccharide Monooxygenase. Curr. Issues Mol. Biol. 2024, 46, 3694-3712. https://doi.org/10.3390/cimb46040230
Li K, Wang Y, Guo X, Wang B. Effects of Lignin-Diverted Reductant with Polyphenol Oxidases on Cellulose Degradation by Wild and Mutant Types of Lytic Polysaccharide Monooxygenase. Current Issues in Molecular Biology. 2024; 46(4):3694-3712. https://doi.org/10.3390/cimb46040230
Chicago/Turabian StyleLi, Kai, Yuan Wang, Xiao Guo, and Bo Wang. 2024. "Effects of Lignin-Diverted Reductant with Polyphenol Oxidases on Cellulose Degradation by Wild and Mutant Types of Lytic Polysaccharide Monooxygenase" Current Issues in Molecular Biology 46, no. 4: 3694-3712. https://doi.org/10.3390/cimb46040230





