Enhancing Yield and Improving Grain Quality in Japonica Rice: Targeted EHD1 Editing via CRISPR-Cas9 in Low-Latitude Adaptation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. Agronomic Trait Measurements
2.3. Plasmid Construction
2.4. Detection of Mutations
2.5. RNA Extraction and RT-PCR
2.6. Photoperiod and Temperature Treatments
2.7. Measurement of Amylose, Gel Consistency, and Alkali Spreading Value
2.8. Statistical Analysis
3. Results
3.1. Early Flowering of Northeastern Japonica Rice at Low Latitudes
3.2. Both ehd1-Ji Yuan Xiang 1 and ehd1-Yi Nong Xiang 12 Display Delayed Heading Date and Good Field Performance
3.3. Temperature Sensitivity of Modified ehd1 Variants
3.4. High Grain Yield and Quality of ehd1 Mutants in Low-Latitude Regions
4. Discussion
4.1. Advancements in Genome Editing for Rice Breeding
4.2. Tailoring Northeastern Japonica Rice for Low-Latitude Cultivation
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Eshed, Y.A.-O.; Lippman, Z.A.-O. Revolutions in agriculture chart a course for targeted breeding of old and new crops. Science 2019, 366, eaax0025. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Kurata, N.; Wei, X.; Wang, Z.-X.; Wang, A.; Zhao, Q.; Zhao, Y.; Liu, K.; Lu, H.; Li, W. A map of rice genome variation reveals the origin of cultivated rice. Nature 2012, 490, 497–501. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Liu, H.; Lin, Y.; Chen, J.; Fu, Y.; Luo, J.; Zhang, Z.; Liang, K.; Chen, S.; Wang, F. In-Frame and Frame-Shift Editing of the Ehd1 Gene to Develop Japonica Rice With Prolonged Basic Vegetative Growth Periods. Front. Plant Sci. 2020, 11, 307. [Google Scholar] [CrossRef] [PubMed]
- Saito, H.; Yuan, Q.; Okumoto, Y.; Doi, K.; Yoshimura, A.; Inoue, H.; Teraishi, M.; Tsukiyama, T.; Tanisaka, T. Multiple alleles at Early flowering 1 locus making variation in the basic vegetative growth period in rice (Oryza sativa L.). Theor. Appl. Genet. 2009, 119, 315–323. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.J.; Jiang, L.; Xu, J.F.; Liu, X.; Liu, S.J.; Zhai, H.Q.; Wan, J.M. The distribution of japonica rice cultivars in the lower region of the Yangtze River valley is determined by its photoperiod-sensitivity and heading date genotypes. J. Integr. Plant Biol. 2009, 51, 922–932. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Q.; Saito, H.; Okumoto, Y.; Inoue, H.; Nishida, H.; Tsukiyama, T.; Teraishi, M.; Tanisaka, T. Identification of a novel gene ef7 conferring an extremely long basic vegetative growth phase in rice. Theor. Appl. Genet. 2009, 119, 675–684. [Google Scholar] [CrossRef] [PubMed]
- Qiu, L.; Wu, Q.; Wang, X.; Han, J.; Zhuang, G.; Wang, H.; Shang, Z.; Tian, W.; Chen, Z.; Lin, Z. Forecasting rice latitude adaptation through a daylength-sensing-based environment adaptation simulator. Nat. Food 2021, 2, 348–362. [Google Scholar] [CrossRef]
- Chen, R.; Deng, Y.; Ding, Y.; Guo, J.; Qiu, J.; Wang, B.; Wang, C.; Xie, Y.; Zhang, Z.; Chen, J.; et al. Rice functional genomics: Decades’ efforts and roads ahead. Sci. China Life Sci. 2022, 65, 33–92. [Google Scholar] [CrossRef] [PubMed]
- Zong, W.; Ren, D.; Huang, M.; Sun, K.; Feng, J.; Zhao, J.; Xiao, D.; Xie, W.; Liu, S.; Zhang, H. Strong photoperiod sensitivity is controlled by cooperation and competition among Hd1, Ghd7 and DTH8 in rice heading. New Phytol. 2021, 229, 1635–1649. [Google Scholar] [CrossRef] [PubMed]
- Rothan, C.; Diouf, I.; Causse, M. Trait discovery and editing in tomato. Plant J. 2019, 97, 73–90. [Google Scholar] [CrossRef]
- Cui, Y.; Hu, X.; Liang, G.; Feng, A.; Wang, F.; Ruan, S.; Dong, G.; Shen, L.; Zhang, B.; Chen, D. Production of novel beneficial alleles of a rice yield-related QTL by CRISPR/Cas9. Plant Biotechnol. J. 2020, 18, 1987. [Google Scholar] [CrossRef]
- Cui, Y.; Zhu, M.; Song, J.; Fan, H.; Xu, X.; Wu, J.; Guo, L.; Wang, J. Expression dynamics of phytochrome genes for the shade-avoidance response in densely direct-seeding rice. Front. Plant Sci. 2022, 13, 1105882. [Google Scholar] [CrossRef] [PubMed]
- Cho, L.-H.; Yoon, J.; Pasriga, R.; An, G. Homodimerization of Ehd1 is required to induce flowering in rice. Plant Physiol. 2016, 170, 2159–2171. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Chen, H.; Ren, D.; Tang, H.; Qiu, R.; Feng, J.; Long, Y.; Niu, B.; Chen, D.; Zhong, T. Genetic interactions between diverged alleles of Early heading date 1 (Ehd1) and Heading date 3a (Hd3a)/RICE FLOWERING LOCUS T1 (RFT 1) control differential heading and contribute to regional adaptation in rice (Oryza sativa). New Phytol. 2015, 208, 936–948. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.L.; Lee, S.; Kim, H.J.; Nam, H.G.; An, G. OsMADS51 is a short-day flowering promoter that functions upstream of Ehd1, OsMADS14, and Hd3a. Plant Physiol. 2007, 145, 1484–1494. [Google Scholar] [CrossRef] [PubMed]
- Andrés, F.; Galbraith, D.W.; Talón, M.; Domingo, C. Analysis of PHOTOPERIOD SENSITIVITY5 sheds light on the role of phytochromes in photoperiodic flowering in rice. Plant Physiol. 2009, 151, 681–690. [Google Scholar] [CrossRef] [PubMed]
- Yano, M.; Katayose, Y.; Ashikari, M.; Yamanouchi, U.; Monna, L.; Fuse, T.; Baba, T.; Yamamoto, K.; Umehara, Y.; Nagamura, Y.; et al. Hd1, a Major Photoperiod Sensitivity Quantitative Trait Locus in Rice, Is Closely Related to the Arabidopsis Flowering Time Gene CONSTANS. Plant Cell 2000, 12, 2473–2483. [Google Scholar] [CrossRef] [PubMed]
- Xue, W.; Xing, Y.; Weng, X.; Zhao, Y.; Tang, W.; Wang, L.; Zhou, H.; Yu, S.; Xu, C.; Li, X. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat. Genet. 2008, 40, 761–767. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.S.; Jeong, D.H.; Lee, D.Y.; Yi, J.; Ryu, C.H.; Kim, S.L.; Jeong, H.J.; Choi, S.C.; Jin, P.; Yang, J. OsCOL4 is a constitutive flowering repressor upstream of Ehd1 and downstream of OsphyB. Plant J. 2010, 63, 18–30. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Xu, J.; Guo, H.; Jiang, L.; Chen, S.; Yu, C.; Zhou, Z.; Hu, P.; Zhai, H.; Wan, J. DTH8 suppresses flowering in rice, influencing plant height and yield potential simultaneously. Plant Physiol. 2010, 153, 1747–1758. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Hu, W.; Shen, G.; Liu, H.; Hu, Y.; Zhou, X.; Liu, T.; Xing, Y. Alternative functions of Hd1 in repressing or promoting heading are determined by Ghd7 status under long-day conditions. Sci. Rep. 2017, 7, 5388. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.S.; Lee, D.Y.; Cho, L.H.; An, G. Rice miR172 induces flowering by suppressing OsIDS1 and SNB, two AP2 genes that negatively regulate expression of Ehd1 and florigens. Rice 2014, 7, 31. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Hua, Y.; Fu, Y.; Li, J.; Liu, Q.; Jiao, X.; Xin, G.; Wang, J.; Wang, X.; Yan, C.; et al. Erratum to: Rapid generation of genetic diversity by multiplex CRISPR/Cas9 genome editing in rice. Sci. China Life Sci. 2019, 62, 1270. [Google Scholar] [CrossRef] [PubMed]
- Hiei, Y.; Komari, T.; Kubo, T. Transformation of rice mediated by Agrobacterium tumefaciens. Plant Mol. Biol. 1997, 35, 205–218. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Xie, X.; Ma, X.; Li, J.; Chen, J.; Liu, Y.G. DSDecode: A Web-Based Tool for Decoding of Sequencing Chromatograms for Genotyping of Targeted Mutations. Mol. Plant 2015, 8, 1431–1433. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Chen, L.; Zhu, Q.; Chen, Y.; Liu, Y.G. Rapid Decoding of Sequence-Specific Nuclease-Induced Heterozygous and Biallelic Mutations by Direct Sequencing of PCR Products. Mol. Plant 2015, 8, 1285–1287. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Zhu, M.; Xu, Z.; Xu, Q. Assessment of the effect of ten heading time genes on reproductive transition and yield components in rice using a CRISPR/Cas9 system. Theor. Appl. Genet. 2019, 132, 1887–1896. [Google Scholar] [CrossRef] [PubMed]
- Anzalone, A.V.; Koblan, L.W.; Liu, D.A.-O. Genome editing with CRISPR-Cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol. 2020, 38, 824–844. [Google Scholar] [CrossRef] [PubMed]
- Son, S.; Park, S.R. Challenges Facing CRISPR/Cas9-Based Genome Editing in Plants. Front. Plant Sci. 2022, 13, 902413. [Google Scholar] [CrossRef] [PubMed]
- Tomlinson, L.; Yang, Y.; Emenecker, R.; Smoker, M.; Taylor, J.; Perkins, S.; Smith, J.; MacLean, D.; Olszewski, N.E.; Jones, J.D. Using CRISPR/Cas9 genome editing in tomato to create a gibberellin-responsive dominant dwarf DELLA allele. Plant Biotechnol. J. 2019, 17, 132–140. [Google Scholar] [CrossRef] [PubMed]
- Macovei, A.; Sevilla, N.R.; Cantos, C.; Jonson, G.B.; Slamet-Loedin, I.; Cermak, T.; Voytas, D.F.; Choi, I.R.; Chadha-Mohanty, P. Novel alleles of rice eIF4G generated by CRISPR/Cas9-targeted mutagenesis confer resistance to Rice tungro spherical virus. Plant Biotechnol. J. 2018, 16, 1918–1927. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Jia, S.; Yobi, A.; Ge, Z.; Sato, S.J.; Zhang, C.; Angelovici, R.; Clemente, T.E.; Holding, D.R. Editing of an alpha-kafirin gene family increases, digestibility and protein quality in sorghum. Plant Physiol. 2018, 177, 1425–1438. [Google Scholar] [CrossRef]
- Liu, X.; Wu, S.; Xu, J.; Sui, C.; Wei, J. Application of CRISPR/Cas9 in plant biology. Acta Pharm. Sin. B 2017, 7, 292–302. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, H.; Botella, J.R.; Zhu, J.K. Generation of new glutinous rice by CRISPR/Cas9-targeted mutagenesis of the Waxy gene in elite rice varieties. J. Integr. Plant Biol. 2018, 60, 369. [Google Scholar] [CrossRef] [PubMed]
- Leng, Y.; Gao, Y.; Chen, L.; Yang, Y.; Huang, L.; Dai, L.; Ren, D.; Xu, Q.; Zhang, Y.; Ponce, K. Using Heading date 1 preponderant alleles from indica cultivars to breed high-yield, high-quality japonica rice varieties for cultivation in south China. Plant Biotechnol. J. 2020, 18, 119–128. [Google Scholar] [CrossRef] [PubMed]
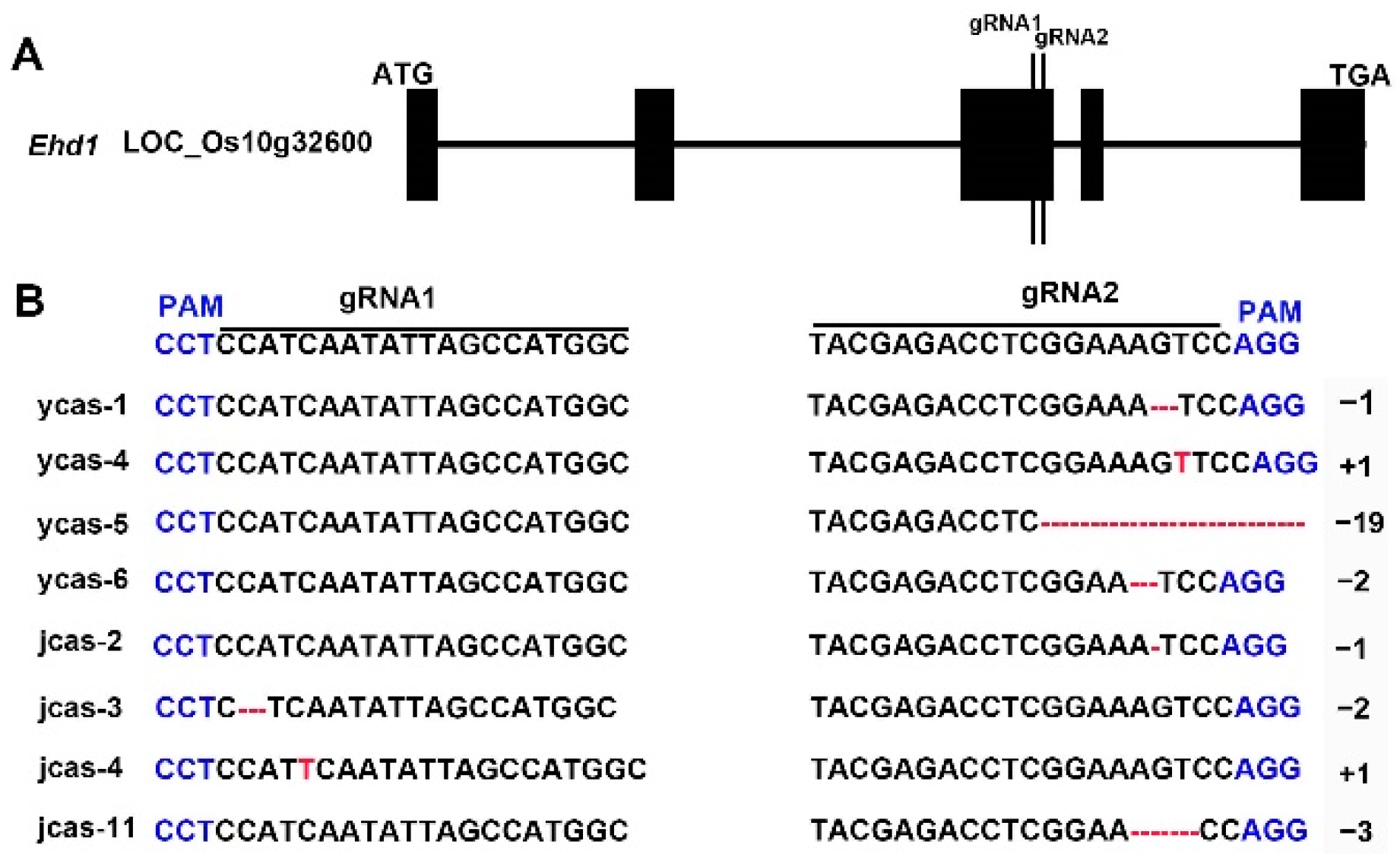


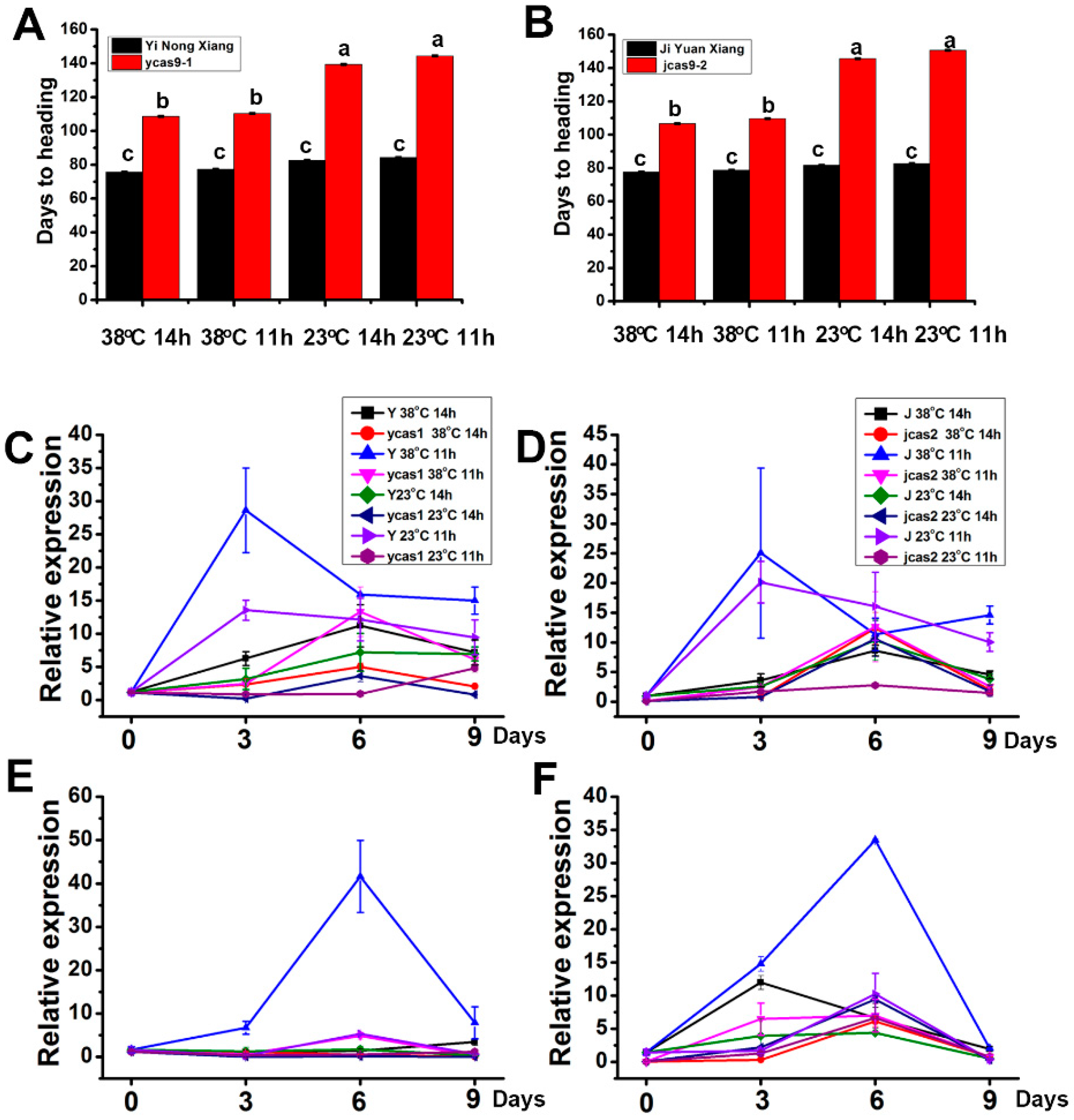
| Rice Line | PH (cm) | PNPP | PL (cm) | PB | SB | SPP | SSR (%) |
|---|---|---|---|---|---|---|---|
| Yinongxiang 12 | 78.3 ± 2.1 a | 7.3 ± 0.5 a | 16.7 ± 0.3 a | 5 ± 1.7 a | 20.7 ± 1.5 a | 76.3 ± 1.5 a | 88.6 ± 1.9 |
| ycas-1 | 102.7 ± 1.5 b | 17.3 ± 0.6 b | 23.2 ± 1.0 b | 11.3 ± 0.6 b | 40.3 ± 2.1 b | 146.3 ± 10.2 b | 88.3 ± 6.7 |
| ycas-4 | 102.3 ± 1.2 b | 19.3 ± 1.5 b | 23.8 ± 0.3 b | 12 ± 1 b | 42.6 ± 4.0 b | 148.7 ± 9.8 b | 91.6 ± 3.2 |
| ycas-5 | 103.7 ± 1.5 b | 16.3 ± 1.5 b | 23.7 ± 0.3 b | 12 ± 1.7 b | 44.3 ± 3.1 b | 145.3 ± 6.0 b | 87.5 ± 2.1 |
| Jiyuanxiang 1 | 83.3 ± 1.2 a | 10.3 ± 1.5 a | 17.6 ± 0.3 a | 10.0 ± 1.7 a | 9.7 ± 1.2 a | 99.3 ± 1.5 a | 83.9 ± 3.5 |
| jcas-2 | 109.3 ± 0.6 b | 18.7 ± 3.1 b | 27 ± 0.1 b | 14.3 ± 1.5 b | 25.3 ± 2.5 b | 227.7 ± 14.3 b | 92.0 ± 1.6 |
| jcas-3 | 110.7 ± 1.5 b | 16.7 ± 1.5 b | 27.1 ± 0.3 b | 14.0 ± 2.6 b | 26.3 ± 1.2 b | 229.3 ± 2.5 b | 89.7 ± 1.7 |
| jcas-4 | 111.7 ± 2.1 b | 19.3 ± 1.5 b | 26.6 ± 0.4 b | 14.6 ± 2.1 b | 22.6 ± 2.9 b | 223.3 ± 5.9 b | 91.2 ± 1.1 |
| Brown Rice Rate (%) | Milled Rice Rate (%) | Head Rice Rate (%) | Grain Length (mm) | Chalkiness Grain Rate (%) | Chalkiness (%) | Amylose (%) | Gel Consistency (mm) | Alkali Spreading Value | |
|---|---|---|---|---|---|---|---|---|---|
| Yi nongxiang | 81.41 ± 0.47 | 72.78 ± 0.72 | 69.85 ± 1.41 | 5.97 ± 0.05 | 52.67 ± 3.51 ** | 12.07 ± 1.66 ** | 17.46 ± 0.24 ** | 55.67 ± 5.13 | 6.60 ± 1.10 |
| ycas9-1 | 82.99 ± 0.279 | 72.59 ± 0.29 | 67.51 ± 0.75 | 6.21 ± 0.01 | 44.00 ± 2.65 | 7.70 ± 0.25 | 15.51 ± 0.44 | 58.00 ± 3.60 | 6.50 ± 0.00 |
| Ji yuanxiang | 82.08 ± 0.44 | 74.38 ± 0.62 | 73.77 ± 0.78 | 4.63 ± 0.02 | 62.33 ± 7.64 ** | 20.03 ± 2.60 ** | 14.57 ± 0.31 | 62.67 ± 0.58 | 6.50 ± 0.00 |
| jcas9-2 | 83.46 ± 0.31 | 74.40 ± 0.41 | 73.36 ± 0.43 | 4.67 ± 0.02 | 32.67 ± 1.53 | 3.70 ± 0.44 | 14.65 ± 0.16 | 62.67 ± 3.06 | 6.70 ± 0.17 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, J.; Tang, L.; Fan, H.; Xu, X.; Peng, X.; Cui, Y.; Wang, J. Enhancing Yield and Improving Grain Quality in Japonica Rice: Targeted EHD1 Editing via CRISPR-Cas9 in Low-Latitude Adaptation. Curr. Issues Mol. Biol. 2024, 46, 3741-3751. https://doi.org/10.3390/cimb46040233
Song J, Tang L, Fan H, Xu X, Peng X, Cui Y, Wang J. Enhancing Yield and Improving Grain Quality in Japonica Rice: Targeted EHD1 Editing via CRISPR-Cas9 in Low-Latitude Adaptation. Current Issues in Molecular Biology. 2024; 46(4):3741-3751. https://doi.org/10.3390/cimb46040233
Chicago/Turabian StyleSong, Jian, Liqun Tang, Honghuan Fan, Xiaozheng Xu, Xinlu Peng, Yongtao Cui, and Jianjun Wang. 2024. "Enhancing Yield and Improving Grain Quality in Japonica Rice: Targeted EHD1 Editing via CRISPR-Cas9 in Low-Latitude Adaptation" Current Issues in Molecular Biology 46, no. 4: 3741-3751. https://doi.org/10.3390/cimb46040233





