Taurine Biosynthesis in a Fish Liver Cell Line (ZFL) Adapted to a Serum-Free Medium
Abstract
:1. Introduction
2. Results
2.1. Taurine Concentrations in L-15 Medium and Fetal Bovine Serum (FBS)
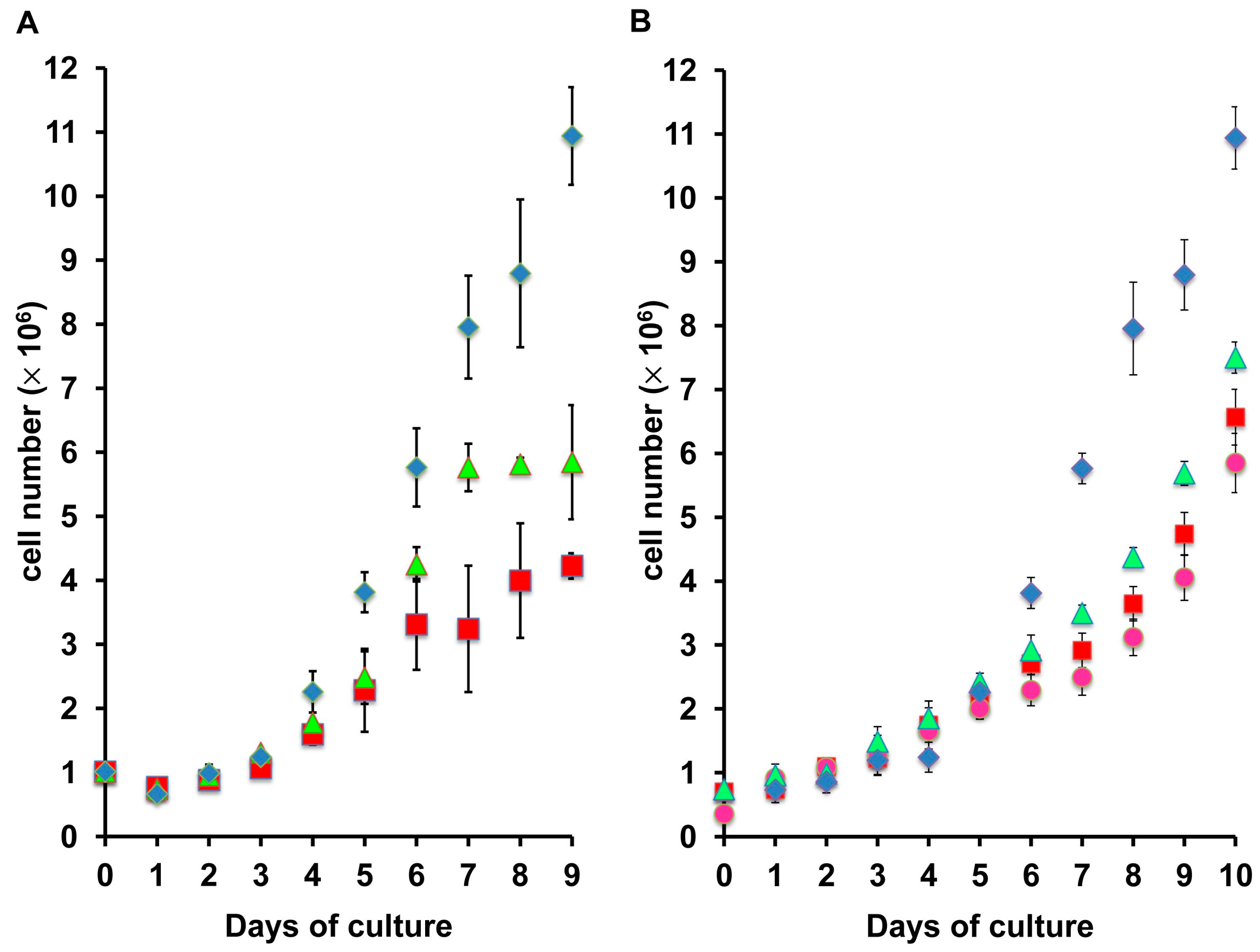
2.2. Adaptation of ZFL Cells to Growth in Serum-Free Medium
2.3. Effect of Taurine Supplementation on Amino Acid Pools in Cells and Medium
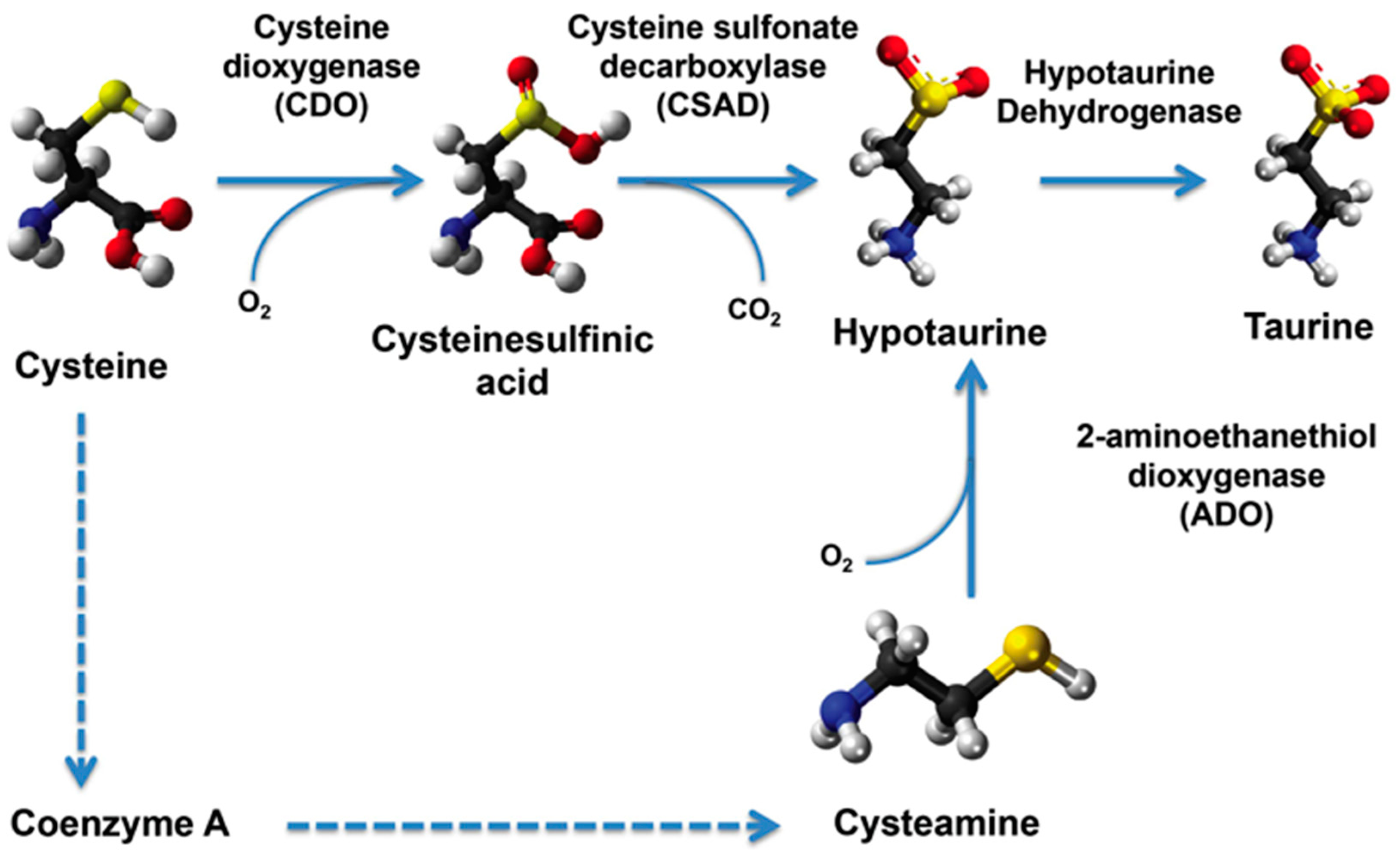
2.4. Transcript Levels of Taurine Transporter and Taurine Biosynthesis Pathway Genes
2.5. Growth Effects of Taurine Are Dependent on Methionine Concentration
3. Discussion
4. Materials and Methods
4.1. Cell Lines and Cell Culture
4.2. Measurement of Medium Amino Acid Concentrations and Cellular Amino Acid Pools
4.3. Measurement of Transcript Levels of the Enzymes of Taurine Biosynthesis
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Ripps, H.; Shen, W. Review: Taurine: A “very essential” amino acid. Mol. Vis. 2012, 18, 2673–2686. [Google Scholar] [PubMed]
- Hofmann, A. Bile Acids: The Good, the Bad, and the Ugly. Physiology 1999, 14, 24–29. [Google Scholar]
- Falany, C.N.; Johnson, M.R.; Barnes, S.; Diasio, R.B. Glycine and taurine conjugation of bile acids by a single enzyme. Molecular cloning and expression of human liver bile acid CoA: Amino acid N-acyltransferase. J. Biol. Chem. 1994, 269, 19375–19379. [Google Scholar] [PubMed]
- Schuller-Levis, G.B.; Park, E. Taurine: New implications for an old amino acid. FEMS Microbiol. Lett. 2003, 226, 195–202. [Google Scholar] [CrossRef]
- Hsu, T.C.; Chiang, S.Y.; Wu, J.H.; Tsai, C.C.; Huang, C.Y.; Chen, Y.C.; Tzang, B.S. Treatment with taurine attenuates hepatic apoptosis in NZB/W F1 mice fed with a high-cholesterol diet. J. Agric. Food Chem. 2008, 56, 9685–9691. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Zhang, Q.; Wang, J.; Liu, F.; Mi, M.; Xu, H.; Chen, F.; Zeng, K. Taurine protects transformed rat retinal ganglion cells from hypoxia-induced apoptosis by preventing mitochondrial dysfunction. Brain Res. 2009, 1279, 131–138. [Google Scholar] [CrossRef] [PubMed]
- Espe, M.; Holen, E. Taurine attenuates apoptosis in primary liver cells isolated from Atlantic salmon (Salmo salar). Br. J. Nutr. 2013, 110, 20–28. [Google Scholar] [CrossRef] [PubMed]
- Lambert, I.H. Regulation of the cellular content of the organic osmolyte taurine in mammalian cells. Neurochem. Res. 2004, 29, 27–63. [Google Scholar] [CrossRef] [PubMed]
- Lambert, I.H.; Kristensen, D.M.; Holm, J.B.; Mortensen, O.H. Physiological role of taurine—From organism to organelle. Acta Physiol. (Oxf.) 2015, 213, 191–212. [Google Scholar] [CrossRef] [PubMed]
- Oja, S.S.; Saransaari, P. Taurine as osmoregulator and neuromodulator in the brain. Metab. Brain Dis. 1996, 11, 153–164. [Google Scholar] [CrossRef] [PubMed]
- Schaffer, S.W.; Azuma, J.; Madura, J.D. Mechanisms underlying taurine-mediated alterations in membrane function. Amino Acids 1995, 8, 231–246. [Google Scholar] [CrossRef] [PubMed]
- Schaffer, S.W.; Jong, C.J.; Ramila, K.C.; Azuma, J. Physiological roles of taurine in heart and muscle. J. Biomed. Sci. 2010, 17 (Suppl. 1), S2. [Google Scholar] [CrossRef] [PubMed]
- Saransaari, P.; Oja, S.S. Enhanced taurine release in cell-damaging conditions in the developing and ageing mouse hippocampus. Neuroscience 1997, 79, 847–854. [Google Scholar] [CrossRef]
- Trenkner, E. Possible role of glutamate with taurine in neuron-glia interaction during cerebellar development. Prog. Clin. Biol. Res. 1990, 351, 133–140. [Google Scholar] [PubMed]
- Hernández-Benítez, R.; Vangipuram, S.D.; Ramos-Mandujano, G.; Lyman, W.D.; Pasantes-Morales, H. Taurine enhances the growth of neural precursors derived from fetal human brain and promotes neuronal specification. Dev. Neurosci. 2013, 35, 40–49. [Google Scholar] [CrossRef] [PubMed]
- Morris, J.G. Idiosyncratic nutrient requirements of cats appear to be diet-induced evolutionary adaptations. Nutr. Res. Rev. 2002, 15, 153–168. [Google Scholar] [CrossRef] [PubMed]
- Verbrugghe, A.; Bakovic, M. Peculiarities of one-carbon metabolism in the strict carnivorous cat and the role in feline hepatic lipidosis. Nutrients 2013, 5, 2811–2835. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Matsunari, H.; Takeuchi, T.; Yokoyama, M.; Murata, Y.; Ishihara, K. Effect of different dietary taurine levels on the conjugated bile acid composition and growth performance of juvenile and fingerling Japanese flounder Paralichthys olivaceus. Aquaculture 2007, 273, 595–601. [Google Scholar] [CrossRef]
- Takagi, S.; Murata, H.; Goto, T.; Hayashi, M.; Hatate, H.; Endo, M.; Yamashito, H.; Ukawa, M. Hemolytic suppression roles of taurine in yellowtail Seriola quinqueradiata fed non-fishmeal diets based on soybean protein. Fish. Sci. 2006, 72, 546–555. [Google Scholar] [CrossRef]
- Omura, Y.; Yoshimura, R. Immunocytochemical localization of taurine in the developing retina of the lefteye flounder Paralichthys olivaceus. Arch. Histol. Cytol. 1999, 62, 441–446. [Google Scholar] [CrossRef] [PubMed]
- Omura, Y.; Inagaki, M. Immunocytochemical localization of taurine in the fish retina under light and dark adaptations. Amino Acids 2000, 19, 593–604. [Google Scholar] [CrossRef] [PubMed]
- Gaylord, T.; Barrows, F.; Teague, A.; Johansen, K.; Overturf, K.; Shepherd, B. Supplementation of taurine and methionine to all-plant protein diets for rainbow trout (Oncorhynchus mykiss). Aquaculture 2007, 269, 514–524. [Google Scholar] [CrossRef]
- Kim, S.; Matsunari, H.; Namura, K.; Tanaka, H.; Yokoyama, M.; Murata, Y.; Ishihara, K. Effect of dietary taurine and lipid contents on conjugated bile acid composition and growth performance of juvenile Japanese flounder Paralichthys olivaceus. Fish. Sci. 2008, 74, 875–881. [Google Scholar] [CrossRef]
- Wang, Q. Dietary sulfur amino acid modulations of taurine biosynthesis in juvenile turbot (Psetta maxima). Aquaculture 2014, 422–423, 141–145. [Google Scholar] [CrossRef]
- Matsunari, H.; Yamamoto, T.; Kim, S.; Goto, T.; Takeuchi, T. Optimum dietary taurine level in casein-based diet for juvenile red sea bream Pagrus major. Fish. Sci. 2008, 74, 347–353. [Google Scholar] [CrossRef]
- Kim, S.; Sasaki, T.; Awa, M.; Inamata, M.; Honryo, T.; Agawa, Y.; Ando, M.; Sawada, Y. Effect of dietary taurine enhancement on growth and development in red sea bream Pagrus major larvae. Aquac. Res. 2016, 47, 1168–1179. [Google Scholar] [CrossRef]
- Lunger, A.; McLean, E.; Gaylord, T.; Kuhn, D.; Craig, S. Taurine supplementation to alternative dietary proteins used in fish meal replacement enhances growth of juvenile cobia (Rachycentron canadum). Aquaculture 2007, 271, 401–410. [Google Scholar] [CrossRef]
- Salze, G.; Craig, S.R.; Smith, B.H.; Smith, E.P.; McLean, E. Morphological development of larval cobia Rachycentron canadum and the influence of dietary taurine supplementation. J. Fish Biol. 2011, 78, 1470–1491. [Google Scholar] [CrossRef] [PubMed]
- Salze, G.; McLean, E.; Craig, S.R. Dietary taurine enhances growth and digestive enzyme activities in larval cobia. Aquaculture 2012, 362–363, 44–49. [Google Scholar] [CrossRef]
- Watson, A.M.; Barrows, F.T.; Place, A.R. Taurine supplementation of plant derived protein and n-3 fatty acids are critical for optimal growth and development of cobia, Rachycentron canadum. Lipids 2013, 48, 899–913. [Google Scholar] [CrossRef] [PubMed]
- Watson, A.M.; Buentello, A.; Place, A.R. Partial replacement of fishmeal, poultry by-product meal and soy concentrate with two non-genetically modified soybean cultivars in diets for juvenile cobia, Rachycentron canadum. Aquaculture 2014, 434, 129–136. [Google Scholar] [CrossRef]
- Espe, M.; Ruohonen, K.; El-Mowafi, A. Effect of taurine supplementation on the metabolism and body lipid-to-protein ratio in juvenile Atlantic salmon (Salmo salar). Aquac. Res. 2012, 271, 349–360. [Google Scholar] [CrossRef]
- Takagi, S.; Murata, H.; Goto, T.; Endo, M.; Yamashita, H.; Ukawa, M. Taurine is an essential nutrient for yellowtail Seriola quinqueradiata fed non-fish meal diets based on soy protein concentrate. Aquaculture 2008, 280, 198–205. [Google Scholar] [CrossRef]
- Kataoka, H.; Ohnishi, N. Occurrence of taurine in plants. Agric. Biol. Chem. 1986, 50, 1887–1888. [Google Scholar]
- Salze, G.P.; Davis, D.A. Taurine: A critical nutrient for future fish feeds. Aquaculture 2015, 437, 215–229. [Google Scholar] [CrossRef]
- O'Flaherty, L.; Stapleton, P.P.; Redmond, H.P.; Bouchier-Hayes, D.J. Intestinal taurine transport: A review. Eur. J. Clin. Investig. 1997, 27, 873–880. [Google Scholar] [CrossRef]
- Tappaz, M.L. Taurine biosynthetic enzymes and taurine transporter: Molecular identification and regulations. Neurochem. Res. 2004, 29, 83–96. [Google Scholar] [CrossRef] [PubMed]
- Brunner, D.; Frank, J.; Appl, H.; Schöffl, H.; Pfaller, W.; Gstraunthaler, G. Serum-free cell culture: The serum-free media interactive online database. Altex 2010, 27, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Van der Valk, J.; Brunner, D.; De Smet, K.; Svenningsen, A.; Honegger, P.; Knudsen, L.E.; Lindl, T.; Noraberg, J.; Price, A.; Scarino, M.L.; et al. Optimization of chemically defined cell culture media—Replacing fetal bovine serum in mammalian in vitro methods. Toxicol. In Vitro 2010, 24, 1053–1063. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.C.; Ding, S.T.; Lee, Y.H.; Wang, Y.C.; Huang, M.F.; Liu, I.H. Taurine homeostasis requires de novo synthesis via cysteine sulfinic acid decarboxylase during zebrafish early embryogenesis. Amino Acids 2013, 44, 615–629. [Google Scholar] [CrossRef] [PubMed]
- Watson, A.M. Taurine: An Indispensable Ingredient in the Development of Sustainable Aquafeeds. Ph.D. Thesis, University of Maryland College Park, Maryland, MD, USA, 2013. [Google Scholar]
- Bjare, U. Serum-free cell culture. Pharmacol. Ther. 1992, 53, 355–374. [Google Scholar] [CrossRef]
- Helmy, M.H.; Ismail, S.S.; Fayed, H.; El-Bassiouni, E.A. Effect of selenium supplementation on the activities of glutathione metabolizing enzymes in human hepatoma Hep G2 cell line. Toxicology 2000, 144, 57–61. [Google Scholar] [CrossRef]
- Karlenius, T.C.; Shah, F.; Yu, W.C.; Hawkes, H.J.; Tinggi, U.; Clarke, F.M.; Tonissen, K.F. The selenium content of cell culture serum influences redox-regulated gene expression. Biotechniques 2011, 50, 295–301. [Google Scholar] [PubMed]
- Pirkmajer, S.; Chibalin, A.V. Serum starvation: Caveat emptor. Am. J. Physiol. Cell Physiol. 2011, 301, C272–C279. [Google Scholar] [CrossRef] [PubMed]
- Bitoun, M.; Tappaz, M. Taurine down-regulates basal and osmolarity-induced gene expression of its transporter, but not the gene expression of its biosynthetic enzymes, in astrocyte primary cultures. J. Neurochem. 2000, 75, 919–924. [Google Scholar] [CrossRef] [PubMed]
- Nutrient Approval is Victory for U.S. Fish Farmers and Seafood Lovers. Available online: http://www.nmfs.noaa.gov/stories/2017/02/taurine-QA.html (accessed on 9 February 2017).
- European Food Safety Authoriy (EFSA). Scientific Opinion on the safety and efficacy of taurine as a feed additive for all animal species: EFSA Panel on Additives and Products or Substances used in Animal Feed (FEEDAP). EFSA J. 2012, 10, 2736. [Google Scholar]
- Rozen, R.; Scriver, C.R. Renal transport of taurine adapts to perturbed taurine homeostasis. Proc. Natl. Acad. Sci. USA 1982, 79, 2101–2105. [Google Scholar] [CrossRef] [PubMed]
- Kozlowski, D.J.; Chen, Z.; Zhuang, L.; Fei, Y.J.; Navarre, S.; Ganapathy, V. Molecular characterization and expression pattern of taurine transporter in zebrafish during embryogenesis. Life Sci. 2008, 82, 1004–1011. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, K.; Toyohara, H.; Sakaguchi, M. A hyperosmotic stress-induced mRNA of carp cell encodes Na+- and Cl−-dependent high affinity taurine transporter. Biochim. Biophys. Acta 2000, 1464, 219–230. [Google Scholar] [CrossRef]
- Zarate, J.; Bradley, T. Molecular cloning and characterization of the taurine transporter of Atlantic salmon. Aquaculture 2007, 273, 209–217. [Google Scholar] [CrossRef]
- Pinto, W.; Rønnestad, I.; Jordal, A.E.; Gomes, A.S.; Dinis, M.T.; Aragão, C. Cloning, tissue and ontogenetic expression of the taurine transporter in the flatfish Senegalese sole (Solea senegalensis). Amino Acids 2012, 42, 1317–1327. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Gen, H.; Xu, W.; Zhou, H. Molecular cloning and characterization of taurine transporter from turbot (Psetta Maxima) and its expression analysis regulated by taurine in vitro. Aquac. Res. 2016, 1–11. [Google Scholar] [CrossRef]
- Eide, M.; Rusten, M.; Male, R.; Jensen, K.H.; Goksøyr, A. A characterization of the ZFL cell line and primary hepatocytes as in vitro liver cell models for the zebrafish (Danio rerio). Aquat. Toxicol. 2014, 147, 7–17. [Google Scholar] [CrossRef] [PubMed]
- Stipanuk, M.H.; Ueki, I.; Dominy, J.E.; Simmons, C.R.; Hirschberger, L.L. Cysteine dioxygenase: A robust system for regulation of cellular cysteine levels. Amino Acids 2009, 37, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Stipanuk, M.H. Sulfur amino acid metabolism: Pathways for production and removal of homocysteine and cysteine. Annu. Rev. Nutr. 2004, 24, 539–577. [Google Scholar] [CrossRef] [PubMed]
- Stipanuk, M.H.; Dominy, J.E.; Lee, J.I.; Coloso, R.M. Mammalian cysteine metabolism: New insights into regulation of cysteine metabolism. J. Nutr. 2006, 136, 1652S–1659S. [Google Scholar] [PubMed]
- Stipanuk, M.H.; Jurkowska, H.; Roman, H.B.; Niewiadomski, J.; Hirschberger, L.L. Insights into taurine synthesis and function based on studies with cysteine dioxygenase (CDO1) knockout mice. Adv. Exp. Med. Biol. 2015, 803, 29–39. [Google Scholar] [PubMed]
- Lee, J.I.; Londono, M.; Hirschberger, L.L.; Stipanuk, M.H. Regulation of cysteine dioxygenase and gamma-glutamylcysteine synthetase is associated with hepatic cysteine level. J. Nutr. Biochem. 2004, 15, 112–122. [Google Scholar] [CrossRef] [PubMed]
- Dominy, J.E.; Hwang, J.; Guo, S.; Hirschberger, L.L.; Zhang, S.; Stipanuk, M.H. Synthesis of amino acid cofactor in cysteine dioxygenase is regulated by substrate and represents a novel post-translational regulation of activity. J. Biol. Chem. 2008, 283, 12188–12201. [Google Scholar] [CrossRef] [PubMed]
- De la Rosa, J.; Stipanuk, M.H. Evidence for a rate-limiting role of cysteinesulfinate decarboxylase activity in taurine biosynthesis in vivo. Comp. Biochem. Physiol. B 1985, 81, 565–571. [Google Scholar] [CrossRef]
- Park, E.; Park, S.Y.; Dobkin, C.; Schuller-Levis, G. Development of a novel cysteine sulfinic Acid decarboxylase knockout mouse: Dietary taurine reduces neonatal mortality. J. Amino Acids 2014, 2014, 346809. [Google Scholar] [CrossRef] [PubMed]
- Park, E.; Park, S.Y.; Dobkin, C.; Schuller-Levis, G. A novel cysteine sulfinic acid decarboxylase knock-out mouse: Comparison between newborn and weanling mice. Adv. Exp. Med. Biol. 2015, 803, 3–16. [Google Scholar] [PubMed]
- Bekaert, M. Reconstruction of Danio rerio metabolic model accounting for subcellular compartmentalisation. PLoS ONE 2016, 7, e49903. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.W.; Hsu, C.C.; Schloss, J.V.; Faiman, M.D.; Wu, E.; Yang, C.Y.; Wu, J.Y. Protein phosphorylation and taurine biosynthesis in vivo and in vitro. J. Neurosci. 1997, 17, 6947–6951. [Google Scholar] [PubMed]
- Takagi, S.; Murata, H.; Goto, T.; Hatate, H.; Endo, M.; Yamashita, H.; Miyatake, H.; Ukawa, M. Role of taurine deficiency in inducing green liver symptom and effect of dietary taurine supplementation in improving growth in juvenile red sea bream Pagrus major fed non-fishmeal diets based on soy protein concentrate. Fish. Sci. 2011, 77, 235–244. [Google Scholar] [CrossRef]
- Johnson, R.B.; Kim, S.K.; Watson, A.M.; Barrows, F.T.; Kroeger, E.L.; Nicklason, P.M.; Goetz, G.W.; Place, A.R. Effects of dietary taurine supplementation on growth, feed efficiency, and nutrient composition of juvenile sablefish (Anoplopoma fimbria) fed plant based feeds. Aquaculture 2015, 445, 79–85. [Google Scholar] [CrossRef]
- Ghosh, C.; Zhou, Y.L.; Collodi, P. Derivation and characterization of a zebrafish liver cell line. Cell Biol. Toxicol. 1994, 10, 167–176. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.; Allagadda, V.; Chibli, H.; Nadeau, J.L.; Mayer, G.D. Comparison of cytotoxicity and expression of metal regulatory genes in zebrafish (Danio rerio) liver cells exposed to cadmium sulfate, zinc sulfate and quantum dots. Metallomics 2013, 5, 1411–1422. [Google Scholar] [CrossRef] [PubMed]
- Cavalcante, D.G.; Da Silva, N.D.; Marcarini, J.C.; Mantovani, M.S.; Marin-Morales, M.A.; Martinez, C.B. Cytotoxic, biochemical and genotoxic effects of biodiesel produced by different routes on ZFL cell line. Toxicol. In Vitro 2014, 28, 1117–1125. [Google Scholar] [CrossRef] [PubMed]
- Pomati, F.; Cotsapas, C.J.; Castiglioni, S.; Zuccato, E.; Calamari, D. Gene expression profiles in zebrafish (Danio rerio) liver cells exposed to a mixture of pharmaceuticals at environmentally relevant concentrations. Chemosphere 2007, 70, 65–73. [Google Scholar] [CrossRef] [PubMed]
- Ruyra, A.; Torrealba, D.; Morera, D.; Tort, L.; MacKenzie, S.; Roher, N. Zebrafish liver (ZFL) cells are able to mount an anti-viral response after stimulation with poly(I:C). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2014, 182C, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, R.D. Establishment and characterization of seven continuous cell lines from freshwater fish. J. Aquat. Anim. Health 1993, 28, 27–34. [Google Scholar] [CrossRef]
- Tang, R.; Dodd, A.; Lai, D.; McNabb, W.C.; Love, D.R. Validation of zebrafish (Danio rerio) reference genes for quantitative real-time RT-PCR normalization. Acta Biochim. Biophys. Sin. 2007, 39, 384–390. [Google Scholar] [CrossRef] [PubMed]


| Source | Taurine Concentration (Mean ± S.D.) |
|---|---|
| L-15 | 11 ± 0.8 μM |
| FBS | 20 ± 4 μM |
| L-15+FBS | 12 ± 1 μM |
| UltraMEM™-ITES | Not detectable |
| At 0 h | After 24 h | ||||
|---|---|---|---|---|---|
| Amino Acid | 0 μM | +0 μM | +12 μM | +160 μM | +2 mM |
| B (D/N) | 68 | 63 ± 1 | 64 ± 2 | 58 ± 4 | 75 ± 5 |
| S | 939 | 774 ± 19 | 778 ± 27 | 682 ± 65 | 817 ± 56 |
| Z (E/Q) | 68 | 91 ± 3 | 94 ± 1 | 88 ± 7 | 120 ± 8 |
| G | 85 | 140 ± 7 | 143 ± 2 | 127 ± 9 | 152 ± 10 |
| H | 774 | 851 ± 32 | 863 ± 17 | 743 ± 69 | 836 ± 49 |
| HYPOTAURINE | 0.00 | 2 ± 0.1 | 3 ± 0.02 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| TAURINE | 0.00 | 0.00 ± 0.0 | 7.4 ± 0.2 | 112 ± 10 | 1,963 ± 131 |
| R | 263 | 245 ± 9 | 244 ± 6 | 219 ± 22 | 236 ± 19 |
| T | 249 | 160 ± 6 | 158 ± 7 | 141 ± 13 | 170 ± 12 |
| A | 105 | 662 ± 5 | 702 ± 7 | 613 ± 56 | 789 ± 37 |
| P | 35 | 25 ± 4 | 19 ± 1 | 17 ± 1 | 21 ± 3 |
| Y | 78 | 73 ± 2 | 72 ± 1 | 65 ± 5 | 76 ± 4 |
| V | 462 | 404 ± 14 | 407 ± 9 | 358 ± 28 | 429 ± 28 |
| METHIONINE | 46 | 19 ± 2 | 16 ± 2 | 15 ± 1 | 14 ± 1 |
| K | 170 | 156 ± 5 | 158 ± 4 | 140 ± 10 | 165 ± 11 |
| I | 210 | 126 ± 4 | 123 ± 3 | 110 ± 8 | 130 ± 9 |
| L | 595 | 471 ± 17 | 473 ± 2 | 416 ± 32 | 506 ± 32 |
| F | 68 | 35 ± 3 | 32 ± 2 | 30 ± 3 | 28 ± 2 |
| Recovery of norleucine (pmol per sample with 50 pmol load) | |||||
| NORLEUCINE * | 50 ± 2 | 50 ± 2 | 51 ± 1 | 51 ± 5 | |
| Medium at 0 h (μM) | Cellular Concentration after 24 h (pmol per 3 × 106 Cells) | ||||
|---|---|---|---|---|---|
| Amino Acid | +0 μM | +12 μM | +160 μM | +2 mM | |
| B (D/N) | 68 | 73 ± 16 | 83 ± 6 | 87 ± 13 | 115 ± 6 |
| S | 939 | 238 ± 9 | 236 ± 11 | 223 ± 27 | 254 ± 16 |
| Z (E/Q) | 68 | 593 ± 22 | 732 ± 40 | 720 ± 100 | 654 ± 33 |
| G | 85 | 94 ± 21 | 119 ± 3 | 112 ± 11 | 135 ± 13 |
| H | 774 | 37 ± 13 | 33 ± 4 | 31 ± 7 | 32 ± 1 |
| HYPOTAURINE | 0.00 | 41 ± 10 | 61 ± 3 | 61 ± 12 | 62 ± 6 |
| TAURINE | 0.00 | 315 ± 63 | 566 ± 34 | 862 ± 34 | 1008 ± 44 |
| R | 263 | 18 ± 5 | ND | ND | ND |
| T | 249 | 54 ± 8 | 59 ± 5 | 57 ± 10 | 66 ± 4 |
| A | 105 | 307 ± 59 | 348 ± 26 | 340 ± 41 | 424 ± 21 |
| P | 35 | 53 ± 8 | 55 ± 10 | 51 ± 10 | 62 ± 2 |
| Y | 78 | 19 ± 2 | 20 ± 2 | 20 ± 3 | 22 ± 1 |
| V | 462 | 86 ± 17 | 93 ± 9 | 91 ± 10 | 103 ± 4 |
| METHIONINE | 46 | 9 ± 1 | 8 ± 0.2 | 8 ± 0.2 | 8 ± 0.20 |
| K | 170 | 21 ± 3 | 19 ± 1 | 20 ± 0.6 | 22 ± 0.1 |
| I | 210 | 33 ± 5 | 34 ± 5 | 34 ± 3 | 37 ± 0.3 |
| L | 595 | 94 ± 23 | 105 ± 9 | 100 ± 9 | 118 ± 6 |
| F | 68 | 24 ± 5 | 25 ± 4 | 23 ± 2 | 23 ± 2 |
| Recovery of norleucine (pmol per sample with 50 pmol load) | |||||
| NORLEUCINE * | 54 ± 3 | 54 ± 3 | 53 ± 2 | 53 ± 2 | |
| Taurine Supplementation | Fold Change in Intracellular Taurine Level | Fold Change in Intracellular Hypotaurine Level | Fold Change in Intracellular Methionine Level |
|---|---|---|---|
| 12 μM | 1.8 | 1.5 | 0.9 |
| 160 μM | 2.8 | 1.5 | 0.9 |
| 2 mM | 3.2 | 1.5 | 0.9 |
| Gene | Forward Primer | Reverse Primer | GenBank Accession Number |
|---|---|---|---|
| D. rerio ADO | 5′-TTACAGACTGCTGGGAAAAA-3′ | 5′-GGCTTGAAACAAGCAAATAA-3′ | NM_001008634.1 |
| D. rerio CDO | 5′-GAACCTGATGGAGTCCTACC-3′ | 5′-AACTTTCCGTTTCCTTCATC-3′ | NM_200741.1 |
| D. rerio CSD | 5′-AGCTGAGATCTCTCCTGGAC-3′ | 5′-TGGTATTGAGGGTTTCAGTG-3′ | NM_001007348 |
| D. rerio TauT | 5′-ATCACCTGTTGGGAGAAACT-3′ | 5′-CAGGTAGTACAAGCCACAGG-3′ | NM_001037661.1 |
| D. rerio L13A | 5′-TCTGGACTGTAAGAGGTATGC-3′ | 5′-AGACGCACAATCTTGAGAGCAG-3′ | NM_212784.1 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, C.-L.; Watson, A.M.; Place, A.R.; Jagus, R. Taurine Biosynthesis in a Fish Liver Cell Line (ZFL) Adapted to a Serum-Free Medium. Mar. Drugs 2017, 15, 147. https://doi.org/10.3390/md15060147
Liu C-L, Watson AM, Place AR, Jagus R. Taurine Biosynthesis in a Fish Liver Cell Line (ZFL) Adapted to a Serum-Free Medium. Marine Drugs. 2017; 15(6):147. https://doi.org/10.3390/md15060147
Chicago/Turabian StyleLiu, Chieh-Lun, Aaron M. Watson, Allen R. Place, and Rosemary Jagus. 2017. "Taurine Biosynthesis in a Fish Liver Cell Line (ZFL) Adapted to a Serum-Free Medium" Marine Drugs 15, no. 6: 147. https://doi.org/10.3390/md15060147
APA StyleLiu, C.-L., Watson, A. M., Place, A. R., & Jagus, R. (2017). Taurine Biosynthesis in a Fish Liver Cell Line (ZFL) Adapted to a Serum-Free Medium. Marine Drugs, 15(6), 147. https://doi.org/10.3390/md15060147






