Interpreting Microbial Biosynthesis in the Genomic Age: Biological and Practical Considerations
Abstract
:1. Introduction
2. Evolution of Biosynthetic Pathways
3. Pathways from Symbiotic and Uncultured Sources
4. Challenges in Biosynthetic Pathway Assembly and Product Prediction
4.1. Capabilities and Limitations of Current Sequencing Technologies
4.2. Metagenomic Binning and Practical Considerations for BGC Analysis
4.3. Strategies to Test and Improve the Accuracy and Contiguity of Assemblies and Pathways
4.4. Challenges, Opportunities, and Parallels to Chemical Analysis in Product Structure Prediction through Bioinformatics
4.5. Successful Examples of Repetitive BGCs Analyzed by De Novo Assembly
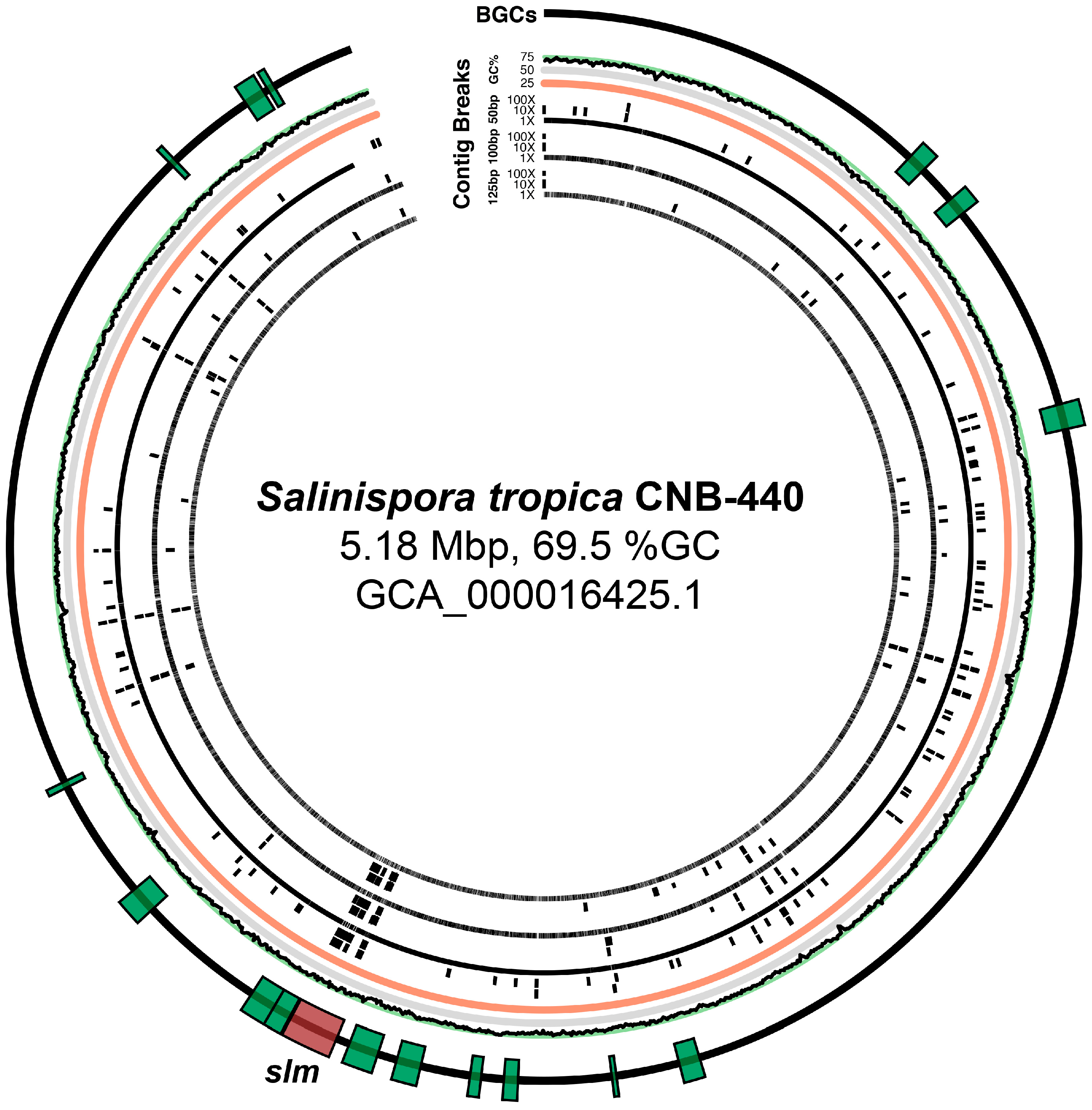
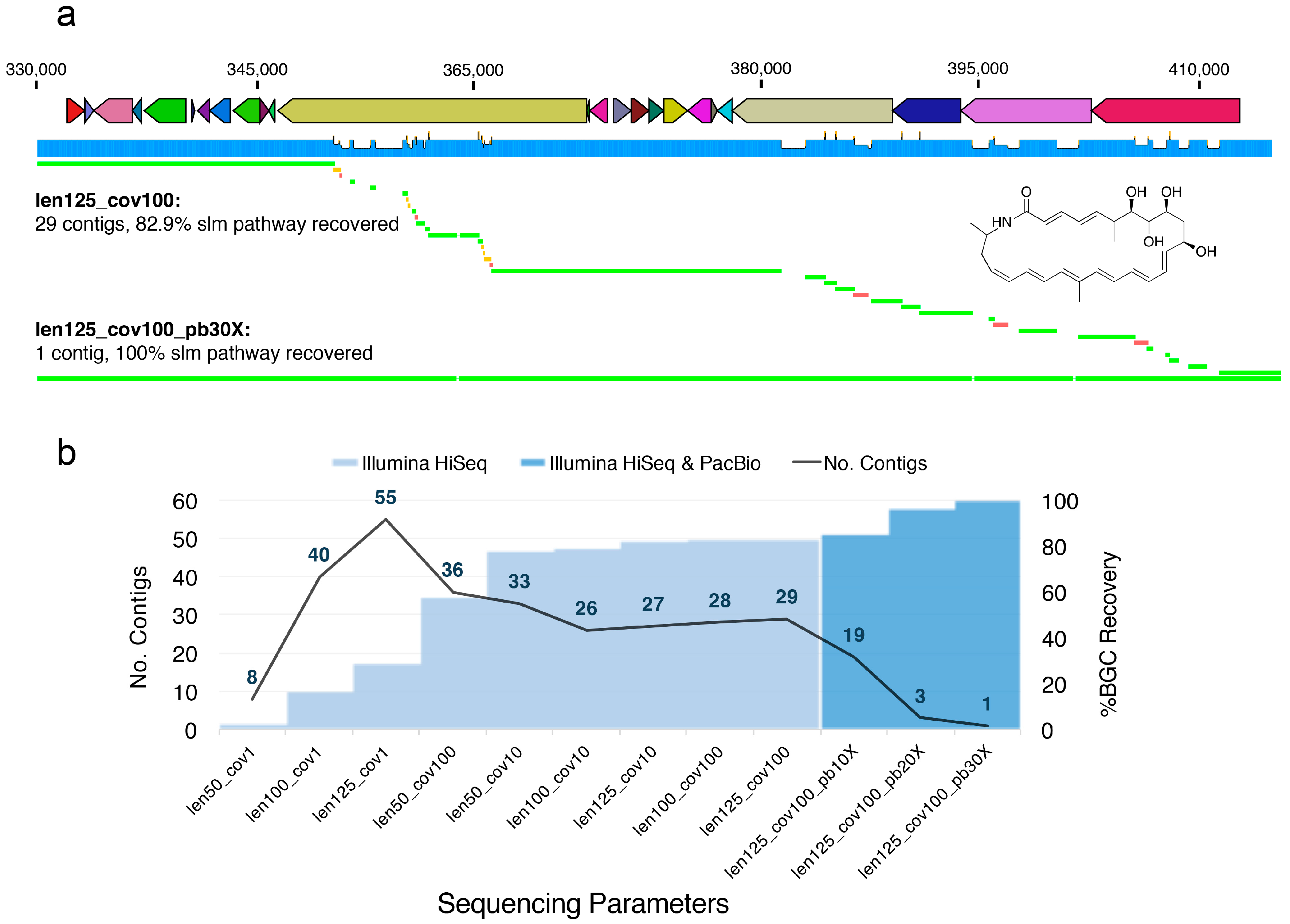
4.6. Analysis of Sequencing Parameters on slm Pathway Fragmentation in Salinispora tropica
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Loman, N.J.; Pallen, M.J. Twenty years of bacterial genome sequencing. Nat. Rev. Microbiol. 2015, 13, 787–794. [Google Scholar] [CrossRef] [PubMed]
- Bashir, A.; Klammer, A.A.; Robins, W.P.; Chin, C.-S.; Webster, D.; Paxinos, E.; Hsu, D.; Ashby, M.; Wang, S.; Peluso, P.; et al. A hybrid approach for the automated finishing of bacterial genomes. Nat. Biotechnol. 2012, 30, 701–707. [Google Scholar] [CrossRef] [PubMed]
- Chin, C.-S.; Alexander, D.H.; Marks, P.; Klammer, A.A.; Drake, J.; Heiner, C.; Clum, A.; Copeland, A.; Huddleston, J.; Eichler, E.E.; et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 2013, 10, 563–569. [Google Scholar] [CrossRef] [PubMed]
- Lewin, G.R.; Carlos, C.; Chevrette, M.G.; Horn, H.A.; McDonald, B.R.; Stankey, R.J.; Fox, B.G.; Currie, C.R. Evolution and ecology of Actinobacteria and their bioenergy applications. Annu. Rev. Microbiol. 2016, 70, 235–254. [Google Scholar] [CrossRef] [PubMed]
- Adnani, N.; Ellis, G.A.; Wyche, T.P.; Bugni, T.S.; Kwan, J.C.; Schmidt, E.W. Emerging trends for stimulating the discovery of natural products. In Natural Products Analysis; John Wiley & Sons: Hoboken, NJ, USA, 2014; pp. 115–161. [Google Scholar]
- Mak, S.; Xu, Y.; Nodwell, J.R. The expression of antibiotic resistance genes in antibiotic-producing bacteria. Mol. Microbiol. 2014, 93, 391–402. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Li, J.; Millán-Aguiñaga, N.; Zhang, J.J.; O’Neill, E.C.; Ugalde, J.A.; Jensen, P.R.; Mantovani, S.M.; Moore, B.S. Identification of thiotetronic acid antibiotic biosynthetic pathways by target-directed genome mining. ACS Chem. Biol. 2015, 10, 2841–2849. [Google Scholar] [CrossRef] [PubMed]
- Hagen, A.; Poust, S.; de Rond, T.; Fortman, J.L.; Katz, L.; Petzold, C.J.; Keasling, J.D. Engineering a polyketide synthase for in vitro production of adipic acid. ACS Synth. Biol. 2016, 5, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Phelan, R.M.; Sekurova, O.N.; Keasling, J.D.; Zotchev, S.B. Engineering terpene biosynthesis in Streptomyces for production of the advanced biofuel precursor bisabolene. ACS Synth. Biol. 2015, 4, 393–399. [Google Scholar] [CrossRef] [PubMed]
- Yamanaka, K.; Reynolds, K.A.; Kersten, R.D.; Ryan, K.S.; Gonzalez, D.J.; Nizet, V.; Dorrestein, P.C.; Moore, B.S. Direct cloning and refactoring of a silent lipopeptide biosynthetic gene cluster yields the antibiotic taromycin A. Proc. Natl. Acad. Sci. USA 2014, 111, 1957–1962. [Google Scholar] [CrossRef] [PubMed]
- Dias, O.; Rocha, M.; Ferreira, E.C.; Rocha, I. Reconstructing genome-scale metabolic models with merlin. Nucleic Acids Res. 2015, 43, 3899–3910. [Google Scholar] [CrossRef] [PubMed]
- Medema, M.H.; Blin, K.; Cimermancic, P.; de Jager, V.; Zakrzewski, P.; Fischbach, M.A.; Weber, T.; Takano, E.; Breitling, R. antiSMASH: Rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 2011, 39, W339–W346. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Wolf, T.; Chevrette, M.G.; Lu, X.; Schwalen, C.J.; Kautsar, S.A.; Suarez Duran, H.G.; de los Santos, E.L.C.; Kim, H.U.; Nave, M.; et al. antiSMASH 4.0—Improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Res. 2017. [Google Scholar] [CrossRef] [PubMed]
- Weimann, A.; Mooren, K.; Frank, J.; Pope, P.B.; Bremges, A.; McHardy, A.C. From genomes to phenotypes: Traitar, the microbial trait analyzer. mSystems 2016, 1, e00101–e00116. [Google Scholar] [CrossRef] [PubMed]
- Bennett, G.M.; Moran, N.A. Heritable symbiosis: The advantages and perils of an evolutionary rabbit hole. Proc. Natl. Acad. Sci. USA 2015, 112, 10169–10176. [Google Scholar] [CrossRef] [PubMed]
- McCutcheon, J.P.; Moran, N.A. Extreme genome reduction in symbiotic bacteria. Nat. Rev. Microbiol. 2012, 10, 13–26. [Google Scholar] [CrossRef] [PubMed]
- Miller, I.J.; Vanee, N.; Fong, S.S.; Lim-Fong, G.E.; Kwan, J.C. Lack of overt genome reduction in the bryostatin-producing bryozoan symbiont “Candidatus Endobugula sertula”. Appl. Environ. Microbiol. 2016, 82, 6573–6583. [Google Scholar] [CrossRef] [PubMed]
- Kwan, J.C.; Schmidt, E.W. Bacterial endosymbiosis in a chordate host: Long-term co-evolution and conservation of secondary metabolism. PLoS ONE 2013, 8, e80822. [Google Scholar] [CrossRef] [PubMed]
- Medema, M.H.; Cimermancic, P.; Sali, A.; Takano, E.; Fischbach, M.A. A systematic computational analysis of biosynthetic gene cluster evolution: Lessons for engineering biosynthesis. PLoS Comput. Biol. 2014, 10, e1004016. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Tyson, G.W.; Eppley, J.M.; DeLong, E.F. Integrated metatranscriptomic and metagenomic analyses of stratified microbial assemblages in the open ocean. ISME J. 2011, 5, 999–1013. [Google Scholar] [CrossRef] [PubMed]
- Haq, I.U.; van Elsas, I.J.D. Metagenomics and metatranscriptomics for the exploration of natural products from soil fungi. In Biosynthesis and Molecular Genetics of Fungal Secondary Metabolites, Volume 2; Zeilinger, S., Martín, J.-F., García-Estrada, C., Eds.; Fungal Biology; Springer: New York, NY, USA, 2015; pp. 67–79. [Google Scholar]
- Clardy, J.; Fischbach, M.A.; Currie, C.R. The natural history of antibiotics. Curr. Biol. 2009, 19, R437–R441. [Google Scholar] [CrossRef] [PubMed]
- Ream, D.C.; Bankapur, A.R.; Friedberg, I. An event-driven approach for studying gene block evolution in bacteria. Bioinformatics 2015, 31, 2075–2083. [Google Scholar] [CrossRef] [PubMed]
- Johnson, S.S.; Chevrette, M.G.; Ehlmann, B.L.; Benison, K.C. Insights from the metagenome of an acid salt lake: The role of biology in an extreme depositional environment. PLoS ONE 2015, 10, e0122869. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.; Oh, D.C. Considerations of the chemical biology of microbial natural products provide an effective drug discovery strategy. Arch. Pharm. Res. 2015, 38, 1591–1605. [Google Scholar] [CrossRef] [PubMed]
- Flórez, L.V.; Biedermann, P.H.W.; Engl, T.; Kaltenpoth, M. Defensive symbioses of animals with prokaryotic and eukaryotic microorganisms. Nat. Prod. Rep. 2015, 32, 904–936. [Google Scholar] [CrossRef] [PubMed]
- Ramadhar, T.R.; Beemelmanns, C.; Currie, C.R.; Clardy, J. Bacterial symbionts in agricultural systems provide a strategic source for antibiotic discovery. J. Antibiot. 2014, 67, 53–58. [Google Scholar] [CrossRef] [PubMed]
- Chevrette, M.G.; Aicheler, F.; Kohlbacher, O.; Currie, C.R.; Medema, M.H. SANDPUMA: Ensemble predictions of nonribosomal peptide chemistry reveals biosynthetic diversity across Actinobacteria. Bioinformatics. in review.
- Calteau, A.; Fewer, D.P.; Latifi, A.; Coursin, T.; Laurent, T.; Jokela, J.; Kerfeld, C.A.; Sivonen, K.; Piel, J.; Gugger, M. Phylum-wide comparative genomics unravel the diversity of secondary metabolism in Cyanobacteria. BMC Genom. 2014, 15, 977. [Google Scholar] [CrossRef] [PubMed]
- Cimermancic, P.; Medema, M.H.; Claesen, J.; Kurita, K.; Wieland Brown, L.C.; Mavrommatis, K.; Pati, A.; Godfrey, P.A.; Koehrsen, M.; Clardy, J.; et al. Insights into secondary metabolism from a global analysis of prokaryotic biosynthetic gene clusters. Cell 2014, 158, 412–421. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Morales, P.; Martínez-Guerrero, C.E. Phylogenomic analysis of natural products biosynthetic gene clusters allows discovery of arseno-organic metabolites in model Streptomycetes. Genome Biol. Evol. 2016, 8, 1906–1916. [Google Scholar] [CrossRef] [PubMed]
- Zucko, J.; Cullum, J.; Hranueli, D.; Long, P.F. Evolutionary dynamics of modular polyketide synthases, with implications for protein design and engineering. J. Antibiot. 2011, 64, 89–92. [Google Scholar] [CrossRef] [PubMed]
- Jenke-Kodama, H.; Dittmann, E. Evolution of metabolic diversity: Insights from microbial polyketide synthases. Phytochemistry 2009, 70, 1858–1866. [Google Scholar] [CrossRef] [PubMed]
- Rounge, T.B.; Rohrlack, T.; Kristensen, T.; Jakobsen, K.S. Recombination and selectional forces in cyanopeptolin NRPS operons from highly similar, but geographically remote Planktothrix strains. BMC Microbiol. 2008, 8, 141. [Google Scholar] [CrossRef] [PubMed]
- Rausch, C.; Hoof, I.; Weber, T.; Wohlleben, W.; Huson, D.H. Phylogenetic analysis of condensation domains in NRPS sheds light on their functional evolution. BMC Evol. Biol. 2007, 7, 78. [Google Scholar] [CrossRef] [PubMed]
- Jenke-Kodama, H.; Börner, T.; Dittmann, E. Natural biocombinatorics in the polyketide synthase genes of the actinobacterium Streptomyces avermitilis. PLoS Comput. Biol. 2006, 2, e132. [Google Scholar] [CrossRef] [PubMed]
- Helfrich, E.J.N.; Piel, J. Biosynthesis of polyketides by trans-AT polyketide synthases. Nat. Prod. Rep. 2016, 33, 231–316. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, D.D.; Melnik, A.V.; Koyama, N.; Lu, X.; Schorn, M.; Fang, J.; Aguinaldo, K.; Lincecum, T.L., Jr.; Ghequire, M.G.K.; Carrion, V.J.; et al. Indexing the Pseudomonas specialized metabolome enabled the discovery of poaeamide B and the bananamides. Nat. Microbiol. 2016, 2, 16197. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; van der Donk, W.A. Ribosomally synthesized and post-translationally modified peptide natural products: New insights into the role of leader and core peptides during biosynthesis. Chemistry 2013, 19, 7662–7677. [Google Scholar] [CrossRef] [PubMed]
- Sardar, D.; Pierce, E.; McIntosh, J.A.; Schmidt, E.W. Recognition sequences and substrate evolution in cyanobactin biosynthesis. ACS Synth. Biol. 2015, 4, 167–176. [Google Scholar] [CrossRef] [PubMed]
- Doroghazi, J.R.; Albright, J.C.; Goering, A.W.; Ju, K.-S.; Haines, R.R.; Tchalukov, K.A.; Labeda, D.P.; Kelleher, N.L.; Metcalf, W.W. A roadmap for natural product discovery based on large-scale genomics and metabolomics. Nat. Chem. Biol. 2014, 10, 963–968. [Google Scholar] [CrossRef] [PubMed]
- Staley, J.T.; Konopka, A. Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Annu. Rev. Microbiol. 1985, 39, 321–346. [Google Scholar] [CrossRef] [PubMed]
- Browne, H.P.; Forster, S.C.; Anonye, B.O.; Kumar, N.; Neville, B.A.; Stares, M.D.; Goulding, D.; Lawley, T.D. Culturing of “unculturable” human microbiota reveals novel taxa and extensive sporulation. Nature 2016, 533, 543–546. [Google Scholar] [CrossRef] [PubMed]
- Ling, L.L.; Schneider, T.; Peoples, A.J.; Spoering, A.L.; Engels, I.; Conlon, B.P.; Mueller, A.; Schäberle, T.F.; Hughes, D.E.; Epstein, S.; et al. A new antibiotic kills pathogens without detectable resistance. Nature 2015, 517, 455–459. [Google Scholar] [CrossRef] [PubMed]
- Lok, C. Mining the microbial dark matter. Nature 2015, 522, 270–273. [Google Scholar] [CrossRef] [PubMed]
- Stewart, E.J. Growing unculturable bacteria. J. Bacteriol. 2012, 194, 4151–4160. [Google Scholar] [CrossRef] [PubMed]
- Yarza, P.; Yilmaz, P.; Pruesse, E.; Glöckner, F.O.; Ludwig, W.; Schleifer, K.-H.; Whitman, W.B.; Euzéby, J.; Amann, R.; Rosselló-Móra, R. Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat. Rev. Microbiol. 2014, 12, 635–645. [Google Scholar] [CrossRef] [PubMed]
- Brown, C.T.; Hug, L.A.; Thomas, B.C.; Sharon, I.; Castelle, C.J.; Singh, A.; Wilkins, M.J.; Wrighton, K.C.; Williams, K.H.; Banfield, J.F. Unusual biology across a group comprising more than 15% of domain Bacteria. Nature 2015, 523, 208–211. [Google Scholar] [CrossRef] [PubMed]
- Rinke, C.; Schwientek, P.; Sczyrba, A.; Ivanova, N.N.; Anderson, I.J.; Cheng, J.-F.; Darling, A.; Malfatti, S.; Swan, B.K.; Gies, E.A.; et al. Insights into the phylogeny and coding potential of microbial dark matter. Nature 2013, 499, 431–437. [Google Scholar] [CrossRef] [PubMed]
- Rath, C.M.; Janto, B.; Earl, J.; Ahmed, A.; Hu, F.Z.; Hiller, L.; Dahlgren, M.; Kreft, R.; Yu, F.; Wolff, J.J.; et al. Meta-omic characterization of the marine invertebrate microbial consortium that produces the chemotherapeutic natural product ET-743. ACS Chem. Biol. 2011, 6, 1244–1256. [Google Scholar] [CrossRef] [PubMed]
- Escobar-Zepeda, A.; Vera-Ponce de León, A.; Sanchez-Flores, A. The road to metagenomics: From microbiology to DNA sequencing technologies and bioinformatics. Front. Genet. 2015, 6, 348. [Google Scholar] [CrossRef] [PubMed]
- Edlund, A.; Loesgen, S.; Fenical, W.; Jensen, P.R. Geographic distribution of secondary metabolite genes in the marine actinomycete Salinispora arenicola. Appl. Environ. Microbiol. 2011, 77, 5916–5925. [Google Scholar] [CrossRef] [PubMed]
- Ziemert, N.; Lechner, A.; Wietz, M.; Millán-Aguiñaga, N.; Chavarria, K.L.; Jensen, P.R. Diversity and evolution of secondary metabolism in the marine actinomycete genus Salinispora. Proc. Natl. Acad. Sci. USA 2014, 111, E1130–E1139. [Google Scholar] [CrossRef] [PubMed]
- Mick, E.; Sorek, R. High-resolution metagenomics. Nat. Biotechnol. 2014, 32, 750–751. [Google Scholar] [CrossRef] [PubMed]
- Compeau, P.E.C.; Pevzner, P.A.; Tesler, G. How to apply de Bruijn graphs to genome assembly. Nat. Biotechnol. 2011, 29, 987–991. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Cleary, B.; Brito, I.L.; Huang, K.; Gevers, D.; Shea, T.; Young, S.; Alm, E.J. Detection of low-abundance bacterial strains in metagenomic datasets by eigengenome partitioning. Nat. Biotechnol. 2015, 33, 1053–1060. [Google Scholar] [CrossRef] [PubMed]
- Nurk, S.; Meleshko, D.; Korobeynikov, A.; Pevzner, P.A. metaSPAdes: A new versatile metagenomic assembler. Genome Res. 2017, 27, 824–834. [Google Scholar] [CrossRef] [PubMed]
- Chang, F.-Y.; Ternei, M.A.; Calle, P.Y.; Brady, S.F. Targeted metagenomics: Finding rare tryptophan dimer natural products in the environment. J. Am. Chem. Soc. 2015, 137, 6044–6052. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.-S.; Brady, S.F. Arixanthomycins A-C: Phylogeny-guided discovery of biologically active eDNA-derived pentangular polyphenols. ACS Chem. Biol. 2014, 9, 1267–1272. [Google Scholar] [CrossRef] [PubMed]
- Freeman, M.F.; Gurgui, C.; Helf, M.J.; Morinaka, B.I.; Uria, A.R.; Oldham, N.J.; Sahl, H.-G.; Matsunaga, S.; Piel, J. Metagenome mining reveals polytheonamides as posttranslationally modified ribosomal peptides. Science 2012, 338, 387–390. [Google Scholar] [CrossRef] [PubMed]
- Schofield, M.M.; Jain, S.; Porat, D.; Dick, G.J.; Sherman, D.H. Identification and analysis of the bacterial endosymbiont specialized for production of the chemotherapeutic natural product ET-743. Environ. Microbiol. 2015, 17, 3964–3975. [Google Scholar] [CrossRef] [PubMed]
- Kwan, J.C.; Donia, M.S.; Han, A.W.; Hirose, E.; Haygood, M.G.; Schmidt, E.W. Genome streamlining and chemical defense in a coral reef symbiosis. Proc. Natl. Acad. Sci. USA 2012, 109, 20655–20660. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, E.W.; Nelson, J.T.; Rasko, D.A.; Sudek, S.; Eisen, J.A.; Haygood, M.G.; Ravel, J. Patellamide A and C biosynthesis by a microcin-like pathway in Prochloron didemni, the cyanobacterial symbiont of Lissoclinum patella. Proc. Natl. Acad. Sci. USA 2005, 102, 7315–7320. [Google Scholar] [CrossRef] [PubMed]
- Nakabachi, A.; Ueoka, R.; Oshima, K.; Teta, R.; Mangoni, A.; Gurgui, M.; Oldham, N.J.; van Echten-Deckert, G.; Okamura, K.; Yamamoto, K.; et al. Defensive bacteriome symbiont with a drastically reduced genome. Curr. Biol. 2013, 23, 1478–1484. [Google Scholar] [CrossRef] [PubMed]
- Kampa, A.; Gagunashvili, A.N.; Gulder, T.A.M.; Morinaka, B.I.; Daolio, C.; Godejohann, M.; Miao, V.P.W.; Piel, J.; Andrésson, Ó.S. Metagenomic natural product discovery in lichen provides evidence for a family of biosynthetic pathways in diverse symbioses. Proc. Natl. Acad. Sci. USA 2013, 110, E3129–E3137. [Google Scholar] [CrossRef] [PubMed]
- Piel, J.; Wen, G.; Platzer, M.; Hui, D. Unprecedented diversity of catalytic domains in the first four modules of the putative pederin polyketide synthase. Chembiochem 2004, 5, 93–98. [Google Scholar] [CrossRef] [PubMed]
- Piel, J. A polyketide synthase-peptide synthetase gene cluster from an uncultured bacterial symbiont of Paederus beetles. Proc. Natl. Acad. Sci. USA 2002, 99, 14002–14007. [Google Scholar] [CrossRef] [PubMed]
- Fisch, K.M.; Gurgui, C.; Heycke, N.; van der Sar, S.A.; Anderson, S.A.; Webb, V.L.; Taudien, S.; Platzer, M.; Rubio, B.K.; Robinson, S.J.; et al. Polyketide assembly lines of uncultivated sponge symbionts from structure-based gene targeting. Nat. Chem. Biol. 2009, 5, 494–501. [Google Scholar] [CrossRef] [PubMed]
- Piel, J.; Hui, D.; Wen, G.; Butzke, D.; Platzer, M.; Fusetani, N.; Matsunaga, S. Antitumor polyketide biosynthesis by an uncultivated bacterial symbiont of the marine sponge Theonella swinhoei. Proc. Natl. Acad. Sci. USA 2004, 101, 16222–16227. [Google Scholar] [CrossRef] [PubMed]
- Donia, M.S.; Cimermancic, P.; Schulze, C.J.; Wieland Brown, L.C.; Martin, J.; Mitreva, M.; Clardy, J.; Linington, R.G.; Fischbach, M.A. A systematic analysis of biosynthetic gene clusters in the human microbiome reveals a common family of antibiotics. Cell 2014, 158, 1402–1414. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.-J.; Chang, F.-Y.; Wyche, T.P.; Backus, K.M.; Acker, T.M.; Funabashi, M.; Taketani, M.; Donia, M.S.; Nayfach, S.; Pollard, K.S.; et al. Discovery of reactive microbiota-derived metabolites that inhibit host proteases. Cell 2017, 168, 517–526. [Google Scholar] [CrossRef] [PubMed]
- Vila-Farres, X.; Chu, J.; Inoyama, D.; Ternei, M.A.; Lemetre, C.; Cohen, L.J.; Cho, W.; Reddy, B.V.B.; Zebroski, H.A.; Freundlich, J.S.; et al. Antimicrobials inspired by nonribosomal peptide synthetase gene clusters. J. Am. Chem. Soc. 2017, 139, 1404–1407. [Google Scholar] [CrossRef] [PubMed]
- Smanski, M.J.; Bhatia, S.; Zhao, D.; Park, Y.; Woodruff, L.B.A.; Giannoukos, G.; Ciulla, D.; Busby, M.; Calderon, J.; Nicol, R.; et al. Functional optimization of gene clusters by combinatorial design and assembly. Nat. Biotechnol. 2014, 32, 1241–1249. [Google Scholar] [CrossRef] [PubMed]
- Smanski, M.J.; Zhou, H.; Claesen, J.; Shen, B.; Fischbach, M.A.; Voigt, C.A. Synthetic biology to access and expand nature’s chemical diversity. Nat. Rev. Microbiol. 2016, 14, 135–149. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, H.A.; Low-Beinart, L.; Obiajulu, J.U.; Brady, S.F. Natural product discovery through improved functional metagenomics in Streptomyces. J. Am. Chem. Soc. 2016, 138, 9341–9344. [Google Scholar] [CrossRef] [PubMed]
- Kelsic, E.D.; Zhao, J.; Vetsigian, K.; Kishony, R. Counteraction of antibiotic production and degradation stabilizes microbial communities. Nature 2015, 521, 516–519. [Google Scholar] [CrossRef] [PubMed]
- Wright, E.S.; Vetsigian, K.H. Inhibitory interactions promote frequent bistability among competing bacteria. Nat. Commun. 2016, 7, 11274. [Google Scholar] [CrossRef] [PubMed]
- Brady, S.F.; Chao, C.J.; Handelsman, J.; Clardy, J. Cloning and heterologous expression of a natural product biosynthetic gene cluster from eDNA. Org. Lett. 2001, 3, 1981–1984. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.Y.; Graziani, E.; Waters, B.; Pan, W.; Li, X.; McDermott, J.; Meurer, G.; Saxena, G.; Andersen, R.J.; Davies, J. Novel natural products from soil DNA libraries in a streptomycete host. Org. Lett. 2000, 2, 2401–2404. [Google Scholar] [CrossRef] [PubMed]
- Bok, J.W.; Ye, R.; Clevenger, K.D.; Mead, D.; Wagner, M.; Krerowicz, A.; Albright, J.C.; Goering, A.W.; Thomas, P.M.; Kelleher, N.L.; et al. Fungal artificial chromosomes for mining of the fungal secondary metabolome. BMC Genom. 2015, 16, 343. [Google Scholar] [CrossRef] [PubMed]
- Craig, J.W.; Cherry, M.A.; Brady, S.F. Long-chain N-acyl amino acid synthases are linked to the putative PEP-CTERM/exosortase protein-sorting system in Gram-negative bacteria. J. Bacteriol. 2011, 193, 5707–5715. [Google Scholar] [CrossRef] [PubMed]
- Cohen, L.J.; Kang, H.-S.; Chu, J.; Huang, Y.-H.; Gordon, E.A.; Reddy, B.V.B.; Ternei, M.A.; Craig, J.W.; Brady, S.F. Functional metagenomic discovery of bacterial effectors in the human microbiome and isolation of commendamide, a GPCR G2A/132 agonist. Proc. Natl. Acad. Sci. USA 2015, 112, E4825–E4834. [Google Scholar] [CrossRef] [PubMed]
- Piel, J. Metabolites from symbiotic bacteria. Nat. Prod. Rep. 2009, 26, 338–362. [Google Scholar] [CrossRef] [PubMed]
- Salvador-Reyes, L.A.; Engene, N.; Paul, V.J.; Luesch, H. Targeted natural products discovery from marine cyanobacteria using combined phylogenetic and mass spectrometric evaluation. J. Nat. Prod. 2015, 78, 486–492. [Google Scholar] [CrossRef] [PubMed]
- Shih, P.M.; Matzke, N.J. Primary endosymbiosis events date to the later Proterozoic with cross-calibrated phylogenetic dating of duplicated ATPase proteins. Proc. Natl. Acad. Sci. USA 2013, 110, 12355–12360. [Google Scholar] [CrossRef] [PubMed]
- Kamel, L.; Keller-Pearson, M.; Roux, C.; Ané, J.-M. Biology and evolution of arbuscular mycorrhizal symbiosis in the light of genomics. New Phytol. 2017, 213, 531–536. [Google Scholar] [CrossRef] [PubMed]
- Smanski, M.J.; Schlatter, D.C.; Kinkel, L.L. Leveraging ecological theory to guide natural product discovery. J. Ind. Microbiol. Biotechnol. 2016, 43, 115–128. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.-W.; Bai, L.; Clade, D.; Hoffmann, D.; Toelzer, S.; Trinh, K.Q.; Xu, J.; Moss, S.J.; Leistner, E.; Floss, H.G. The biosynthetic gene cluster of the maytansinoid antitumor agent ansamitocin from Actinosynnema pretiosum. Proc. Natl. Acad. Sci. USA 2002, 99, 7968–7973. [Google Scholar] [CrossRef] [PubMed]
- Piel, J.; Höfer, I.; Hui, D. Evidence for a symbiosis island involved in horizontal acquisition of pederin biosynthetic capabilities by the bacterial symbiont of Paederus fuscipes beetles. J. Bacteriol. 2004, 186, 1280–1286. [Google Scholar] [CrossRef] [PubMed]
- Moran, N.A.; Munson, M.A.; Baumann, P.; Ishikawa, H. A molecular clock in endosymbiotic bacteria is calibrated using the insect hosts. Proc. R. Soc. Lond. B Biol. Sci. 1993, 253, 167–171. [Google Scholar] [CrossRef]
- Tamas, I.; Klasson, L.; Canbäck, B.; Näslund, A.K.; Eriksson, A.-S.; Wernegreen, J.J.; Sandström, J.P.; Moran, N.A.; Andersson, S.G.E. 50 million years of genomic stasis in endosymbiotic bacteria. Science 2002, 296, 2376–2379. [Google Scholar] [CrossRef] [PubMed]
- Trindade-Silva, A.E.; Lim-Fong, G.E.; Sharp, K.H.; Haygood, M.G. Bryostatins: Biological context and biotechnological prospects. Curr. Opin. Biotechnol. 2010, 21, 834–842. [Google Scholar] [CrossRef] [PubMed]
- Lim-Fong, G.E.; Regali, L.A.; Haygood, M.G. Evolutionary relationships of “Candidatus endobugula” bacterial symbionts and their Bugula bryozoan hosts. Appl. Environ. Microbiol. 2008, 74, 3605–3609. [Google Scholar] [CrossRef] [PubMed]
- Fehlauer-Ale, K.H.; Mackie, J.A.; Lim-Fong, G.E.; Ale, E.; Pie, M.R.; Waeschenbach, A. Cryptic species in the cosmopolitan Bugula neritina complex (Bryozoa, Cheilostomata). Zool. Scr. 2014, 43, 193–205. [Google Scholar] [CrossRef]
- Hildebrand, M.; Waggoner, L.E.; Liu, H.; Sudek, S.; Allen, S.; Anderson, C.; Sherman, D.H.; Haygood, M. bryA: An unusual modular polyketide synthase gene from the uncultivated bacterial symbiont of the marine bryozoan Bugula neritina. Chem. Biol. 2004, 11, 1543–1552. [Google Scholar] [CrossRef] [PubMed]
- Sudek, S.; Lopanik, N.B.; Waggoner, L.E.; Hildebrand, M.; Anderson, C.; Liu, H.; Patel, A.; Sherman, D.H.; Haygood, M.G. Identification of the putative bryostatin polyketide synthase gene cluster from “Candidatus Endobugula sertula”, the uncultivated microbial symbiont of the marine bryozoan Bugula neritina. J. Nat. Prod. 2007, 70, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Sit, C.S.; Ruzzini, A.C.; Van Arnam, E.B.; Ramadhar, T.R.; Currie, C.R.; Clardy, J. Variable genetic architectures produce virtually identical molecules in bacterial symbionts of fungus-growing ants. Proc. Natl. Acad. Sci. USA 2015, 112, 13150–13154. [Google Scholar] [CrossRef] [PubMed]
- Linneman, J.; Paulus, D.; Lim-Fong, G.; Lopanik, N.B. Latitudinal variation of a defensive symbiosis in the Bugula neritina (Bryozoa) sibling species complex. PLoS ONE 2014, 9, e108783. [Google Scholar] [CrossRef] [PubMed]
- Goodwin, S.; McPherson, J.D.; McCombie, W.R. Coming of age: Ten years of next-generation sequencing technologies. Nat. Rev. Genet. 2016, 17, 333–351. [Google Scholar] [CrossRef] [PubMed]
- Frank, J.A.; Pan, Y.; Tooming-Klunderud, A.; Eijsink, V.G.H.; McHardy, A.C.; Nederbragt, A.J.; Pope, P.B. Improved metagenome assemblies and taxonomic binning using long-read circular consensus sequence data. Sci. Rep. 2016, 6, 25373. [Google Scholar] [CrossRef] [PubMed]
- Siegl, A.; Kamke, J.; Hochmuth, T.; Piel, J.; Richter, M.; Liang, C.; Dandekar, T.; Hentschel, U. Single-cell genomics reveals the lifestyle of Poribacteria, a candidate phylum symbiotically associated with marine sponges. ISME J. 2011, 5, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Piel, J. Approaches to capturing and designing biologically active small molecules produced by uncultured microbes. Annu. Rev. Microbiol. 2011, 65, 431–453. [Google Scholar] [CrossRef] [PubMed]
- Lasken, R.S. Single-cell genomic sequencing using Multiple Displacement Amplification. Curr. Opin. Microbiol. 2007, 10, 510–516. [Google Scholar] [CrossRef] [PubMed]
- Blainey, P.C.; Quake, S.R. Digital MDA for enumeration of total nucleic acid contamination. Nucleic Acids Res. 2011, 39, e19. [Google Scholar] [CrossRef] [PubMed]
- Gawad, C.; Koh, W.; Quake, S.R. Single-cell genome sequencing: Current state of the science. Nat. Rev. Genet. 2016, 17, 175–188. [Google Scholar] [CrossRef] [PubMed]
- Pope, P.B.; Smith, W.; Denman, S.E.; Tringe, S.G.; Barry, K.; Hugenholtz, P.; McSweeney, C.S.; McHardy, A.C.; Morrison, M. Isolation of Succinivibrionaceae implicated in low methane emissions from Tammar wallabies. Science 2011, 333, 646–648. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Chng, K.R.; Wilm, A.; Zhao, S.; Yang, K.-L.; Nagarajan, N.; He, J. Genomic characterization of three unique Dehalococcoides that respire on persistent polychlorinated biphenyls. Proc. Natl. Acad. Sci. USA 2014, 111, 12103–12108. [Google Scholar] [CrossRef] [PubMed]
- Cuív, P.Ó.; Smith, W.J.; Pottenger, S.; Burman, S.; Shanahan, E.R.; Morrison, M. Isolation of genetically tractable most-wanted bacteria by metaparental mating. Sci. Rep. 2015, 5, 13282. [Google Scholar] [CrossRef] [PubMed]
- Tyson, G.W.; Lo, I.; Baker, B.J.; Allen, E.E.; Hugenholtz, P.; Banfield, J.F. Genome-directed isolation of the key nitrogen fixer Leptospirillum ferrodiazotrophum sp. nov. from an acidophilic microbial community. Appl. Environ. Microbiol. 2005, 71, 6319–6324. [Google Scholar] [CrossRef] [PubMed]
- Omsland, A.; Cockrell, D.C.; Howe, D.; Fischer, E.R.; Virtaneva, K.; Sturdevant, D.E.; Porcella, S.F.; Heinzen, R.A. Host cell-free growth of the Q fever bacterium Coxiella burnetii. Proc. Natl. Acad. Sci. USA 2009, 106, 4430–4434. [Google Scholar] [CrossRef] [PubMed]
- Awan, A.R.; Shaw, W.M.; Ellis, T. Biosynthesis of therapeutic natural products using synthetic biology. Adv. Drug Deliv. Rev. 2016, 105, 96–106. [Google Scholar] [CrossRef] [PubMed]
- Owen, J.G.; Reddy, B.V.B.; Ternei, M.A.; Charlop-Powers, Z.; Calle, P.Y.; Kim, J.H.; Brady, S.F. Mapping gene clusters within arrayed metagenomic libraries to expand the structural diversity of biomedically relevant natural products. Proc. Natl. Acad. Sci. USA 2013, 110, 11797–11802. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.Y.; Chen, T.; Fuhrman, J.A.; Sun, F. COCACOLA: Binning metagenomic contigs using sequence Composition, read CoverAge, CO-alignment and paired-end read LinkAge. Bioinformatics 2016, 33, 791–798. [Google Scholar] [CrossRef] [PubMed]
- Chatterji, S.; Yamazaki, I.; Bai, Z.; Eisen, J.A. CompostBin: A DNA composition-based algorithm for binning environmental shotgun reads. In Research in Computational Molecular Biology; Springer: Berlin/Heidelberg, Germany, 2008; pp. 17–28. [Google Scholar]
- Alneberg, J.; Bjarnason, B.S.; de Bruijn, I.; Schirmer, M.; Quick, J.; Ijaz, U.Z.; Lahti, L.; Loman, N.J.; Andersson, A.F.; Quince, C. Binning metagenomic contigs by coverage and composition. Nat. Methods 2014, 11, 1144–1146. [Google Scholar] [CrossRef] [PubMed]
- Sieber, C.M.K.; Probst, A.J.; Sharrar, A.; Thomas, B.C.; Hess, M.; Tringe, S.G.; Banfield, J.F. Recovery of genomes from metagenomes via a dereplication, aggregation, and scoring strategy. bioRxiv 2017, 107789. [Google Scholar] [CrossRef]
- Imelfort, M.; Parks, D.; Woodcroft, B.J.; Dennis, P.; Hugenholtz, P.; Tyson, G.W. GroopM: An automated tool for the recovery of population genomes from related metagenomes. PeerJ 2014, 2, e603. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.-W.; Simmons, B.A.; Singer, S.W. MaxBin 2.0: An automated binning algorithm to recover genomes from multiple metagenomic datasets. Bioinformatics 2016, 32, 605–607. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Hu, H.; Li, X. MBBC: An efficient approach for metagenomic binning based on clustering. BMC Bioinform. 2015, 16, 36. [Google Scholar] [CrossRef] [PubMed]
- Kang, D.D.; Froula, J.; Egan, R.; Wang, Z. MetaBAT, an efficient tool for accurately reconstructing single genomes from complex microbial communities. PeerJ 2015, 3, e1165. [Google Scholar] [CrossRef] [PubMed]
- Strous, M.; Kraft, B.; Bisdorf, R.; Tegetmeyer, H.E. The binning of metagenomic contigs for microbial physiology of mixed cultures. Front. Microbiol. 2012, 3, 410. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Leung, H.C.M.; Yiu, S.M.; Chin, F.Y.L. MetaCluster 5.0: A two-round binning approach for metagenomic data for low-abundance species in a noisy sample. Bioinformatics 2012, 28, i356–i362. [Google Scholar] [CrossRef] [PubMed]
- Albertsen, M.; Hugenholtz, P.; Skarshewski, A.; Nielsen, K.L.; Tyson, G.W.; Nielsen, P.H. Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nat. Biotechnol. 2013, 31, 533–538. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.-H.; Liao, Y.-C. Accurate binning of metagenomic contigs via automated clustering sequences using information of genomic signatures and marker genes. Sci. Rep. 2016, 6, 24175. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, M.H.; Ghosh, T.S.; Singh, N.K.; Mande, S.S. SPHINX—An algorithm for taxonomic binning of metagenomic sequences. Bioinformatics 2011, 27, 22–30. [Google Scholar] [CrossRef] [PubMed]
- Kelley, D.R.; Salzberg, S.L. Clustering metagenomic sequences with interpolated Markov models. BMC Bioinform. 2010, 11, 544. [Google Scholar] [CrossRef] [PubMed]
- Ultsch, A.; Mörchen, F. ESOM-Maps: Tools for Clustering, Visualization, and Classification with Emergent SOM; Technical Report No. 46; Deptartment of Mathematics and Computer Science, University of Marburg: Marburg, Germany, 2005. [Google Scholar]
- Dick, G.J.; Andersson, A.F.; Baker, B.J.; Simmons, S.L.; Thomas, B.C.; Yelton, A.P.; Banfield, J.F. Community-wide analysis of microbial genome sequence signatures. Genome Biol. 2009, 10, R85. [Google Scholar] [CrossRef] [PubMed]
- Laczny, C.C.; Sternal, T.; Plugaru, V.; Gawron, P.; Atashpendar, A.; Margossian, H.H.; Coronado, S.; van der Maaten, L.; Vlassis, N.; Wilmes, P. VizBin—An application for reference-independent visualization and human-augmented binning of metagenomic data. Microbiome 2015, 3, 1. [Google Scholar] [CrossRef] [PubMed]
- Saeed, I.; Tang, S.-L.; Halgamuge, S.K. Unsupervised discovery of microbial population structure within metagenomes using nucleotide base composition. Nucleic Acids Res. 2012, 40, e34. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, H.B.; Almeida, M.; Juncker, A.S.; Rasmussen, S.; Li, J.; Sunagawa, S.; Plichta, D.R.; Gautier, L.; Pedersen, A.G.; Le Chatelier, E.; et al. MetaHIT Consortium Identification and assembly of genomes and genetic elements in complex metagenomic samples without using reference genomes. Nat. Biotechnol. 2014, 32, 822–828. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Leung, H.; Yiu, S.; Chin, F. MetaCluster-TA: Taxonomic annotation for metagenomic data based on assembly-assisted binning. BMC Genom. 2014, 15 (Suppl. 1), S12. [Google Scholar] [CrossRef] [PubMed]
- Sedlar, K.; Kupkova, K.; Provaznik, I. Bioinformatics strategies for taxonomy independent binning and visualization of sequences in shotgun metagenomics. Comput. Struct. Biotechnol. J. 2017, 15, 48–55. [Google Scholar] [CrossRef] [PubMed]
- Cheng, T.Y.; Sueoka, N. Heterogeneity of DNA in density and base composition. Science 1963, 141, 1194–1196. [Google Scholar] [CrossRef] [PubMed]
- Teeling, H.; Meyerdierks, A.; Bauer, M.; Amann, R.; Glöckner, F.O. Application of tetranucleotide frequencies for the assignment of genomic fragments. Environ. Microbiol. 2004, 6, 938–947. [Google Scholar] [CrossRef] [PubMed]
- Laczny, C.C.; Pinel, N.; Vlassis, N.; Wilmes, P. Alignment-free visualization of metagenomic data by nonlinear dimension reduction. Sci. Rep. 2014, 4, 4516. [Google Scholar] [CrossRef] [PubMed]
- Medini, D.; Donati, C.; Tettelin, H.; Masignani, V.; Rappuoli, R. The microbial pan-genome. Curr. Opin. Genet. Dev. 2005, 15, 589–594. [Google Scholar] [CrossRef] [PubMed]
- Tettelin, H.; Riley, D.; Cattuto, C.; Medini, D. Comparative genomics: The bacterial pan-genome. Curr. Opin. Microbiol. 2008, 11, 472–477. [Google Scholar] [CrossRef] [PubMed]
- Miller, I.J.; Weyna, T.R.; Fong, S.S.; Lim-Fong, G.E.; Kwan, J.C. Single sample resolution of rare microbial dark matter in a marine invertebrate metagenome. Sci. Rep. 2016, 6, 34362. [Google Scholar] [CrossRef] [PubMed]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef] [PubMed]
- Mikheenko, A.; Saveliev, V.; Gurevich, A. MetaQUAST: Evaluation of metagenome assemblies. Bioinformatics 2016, 32, 1088–1090. [Google Scholar] [CrossRef] [PubMed]
- Hunt, M.; Kikuchi, T.; Sanders, M.; Newbold, C.; Berriman, M.; Otto, T.D. REAPR: A universal tool for genome assembly evaluation. Genome Biol. 2013, 14, R47. [Google Scholar] [CrossRef] [PubMed]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef] [PubMed]
- Thomas, T.; Gilbert, J.; Meyer, F. Metagenomics—A guide from sampling to data analysis. Microb. Inform. Exp. 2012, 2, 3. [Google Scholar] [CrossRef] [PubMed]
- Sangwan, N.; Xia, F.; Gilbert, J.A. Recovering complete and draft population genomes from metagenome datasets. Microbiome 2016, 4, 8. [Google Scholar] [CrossRef] [PubMed]
- Ashton, P.M.; Nair, S.; Dallman, T.; Rubino, S.; Rabsch, W.; Mwaigwisya, S.; Wain, J.; O’Grady, J. MinION nanopore sequencing identifies the position and structure of a bacterial antibiotic resistance island. Nat. Biotechnol. 2015, 33, 296–300. [Google Scholar] [CrossRef] [PubMed]
- Sharon, I.; Kertesz, M.; Hug, L.A.; Pushkarev, D.; Blauwkamp, T.A.; Castelle, C.J.; Amirebrahimi, M.; Thomas, B.C.; Burstein, D.; Tringe, S.G.; et al. Accurate, multi-kb reads resolve complex populations and detect rare microorganisms. Genome Res. 2015, 25, 534–543. [Google Scholar] [CrossRef] [PubMed]
- Burton, J.N.; Liachko, I.; Dunham, M.J.; Shendure, J. Species-level deconvolution of metagenome assemblies with Hi-C-based contact probability maps. G3 2014, 4, 1339–1346. [Google Scholar] [CrossRef] [PubMed]
- Marbouty, M.; Baudry, L.; Cournac, A.; Koszul, R. Scaffolding bacterial genomes and probing host-virus interactions in gut microbiome by proximity ligation (chromosome capture) assay. Sci. Adv. 2017, 3, e1602105. [Google Scholar] [CrossRef] [PubMed]
- Beitel, C.W.; Froenicke, L.; Lang, J.M.; Korf, I.F.; Michelmore, R.W.; Eisen, J.A.; Darling, A.E. Strain- and plasmid-level deconvolution of a synthetic metagenome by sequencing proximity ligation products. PeerJ 2014, 2, e415. [Google Scholar] [CrossRef] [PubMed]
- Flot, J.-F.; Marie-Nelly, H.; Koszul, R. Contact genomics: Scaffolding and phasing (meta)genomes using chromosome 3D physical signatures. FEBS Lett. 2015, 589, 2966–2974. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Darling, A. Metagenomic Chromosome Conformation Capture (3C): Techniques, applications, and challenges. F1000Research 2015, 4, 1377. [Google Scholar] [CrossRef] [PubMed]
- Marbouty, M.; Cournac, A.; Flot, J.-F.; Marie-Nelly, H.; Mozziconacci, J.; Koszul, R. Metagenomic chromosome conformation capture (meta3C) unveils the diversity of chromosome organization in microorganisms. eLife 2014, 3, e03318. [Google Scholar] [CrossRef] [PubMed]
- Staunton, J.; Weissman, K.J. Polyketide biosynthesis: A millennium review. Nat. Prod. Rep. 2001, 18, 380–416. [Google Scholar] [CrossRef] [PubMed]
- Kamra, P.; Gokhale, R.S.; Mohanty, D. SEARCHGTr: A program for analysis of glycosyltransferases involved in glycosylation of secondary metabolites. Nucleic Acids Res. 2005, 33, W220–W225. [Google Scholar] [CrossRef] [PubMed]
- Hertweck, C. The biosynthetic logic of polyketide diversity. Angew. Chem. Int. Ed. Engl. 2009, 48, 4688–4716. [Google Scholar] [CrossRef] [PubMed]
- Strieker, M.; Tanović, A.; Marahiel, M.A. Nonribosomal peptide synthetases: Structures and dynamics. Curr. Opin. Struct. Biol. 2010, 20, 234–240. [Google Scholar] [CrossRef] [PubMed]
- Yadav, G.; Gokhale, R.S.; Mohanty, D. Computational approach for prediction of domain organization and substrate specificity of modular polyketide synthases. J. Mol. Biol. 2003, 328, 335–363. [Google Scholar] [CrossRef]
- Irschik, H.; Kopp, M.; Weissman, K.J.; Buntin, K.; Piel, J.; Müller, R. Analysis of the sorangicin gene cluster reinforces the utility of a combined phylogenetic/retrobiosynthetic analysis for deciphering natural product assembly by trans-AT PKS. Chembiochem 2010, 11, 1840–1849. [Google Scholar] [CrossRef] [PubMed]
- Jensen, K.; Niederkrüger, H.; Zimmermann, K.; Vagstad, A.L.; Moldenhauer, J.; Brendel, N.; Frank, S.; Pöplau, P.; Kohlhaas, C.; Townsend, C.A.; et al. Polyketide proofreading by an acyltransferase-like enzyme. Chem. Biol. 2012, 19, 329–339. [Google Scholar] [CrossRef] [PubMed]
- Challis, G.L.; Ravel, J.; Townsend, C.A. Predictive, structure-based model of amino acid recognition by nonribosomal peptide synthetase adenylation domains. Chem. Biol. 2000, 7, 211–224. [Google Scholar] [CrossRef]
- Piel, J. Biosynthesis of polyketides by trans-AT polyketide synthases. Nat. Prod. Rep. 2010, 27, 996–1047. [Google Scholar] [CrossRef] [PubMed]
- Sharp, K.H.; Davidson, S.K.; Haygood, M.G. Localization of “Candidatus Endobugula sertula” and the bryostatins throughout the life cycle of the bryozoan Bugula neritina. ISME J. 2007, 1, 693–702. [Google Scholar] [CrossRef] [PubMed]
- Udwary, D.W.; Zeigler, L.; Asolkar, R.N.; Singan, V.; Lapidus, A.; Fenical, W.; Jensen, P.R.; Moore, B.S. Genome sequencing reveals complex secondary metabolome in the marine actinomycete Salinispora tropica. Proc. Natl. Acad. Sci. USA 2007, 104, 10376–10381. [Google Scholar] [CrossRef] [PubMed]
- Lane, A.L.; Moore, B.S. A sea of biosynthesis: Marine natural products meet the molecular age. Nat. Prod. Rep. 2011, 28, 411–428. [Google Scholar] [CrossRef] [PubMed]
- Matsumori, N.; Kaneno, D.; Murata, M.; Nakamura, H.; Tachibana, K. Stereochemical determination of acyclic structures based on carbon-proton spin-coupling constants. A method of configuration analysis for natural products. J. Org. Chem. 1999, 64, 866–876. [Google Scholar] [CrossRef] [PubMed]
- Lei, H.; Yan, J.; Yu, J.; Liu, Y.; Wang, Z.; Xu, Z.; Ye, T. Total synthesis and stereochemical reassignment of mandelalide A. Angew. Chem. Int. Ed. Engl. 2014, 53, 6533–6537. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Li, L.; Myers, J.R.; Marth, G.T. ART: A next-generation sequencing read simulator. Bioinformatics 2012, 28, 593–594. [Google Scholar] [CrossRef] [PubMed]
- Ono, Y.; Asai, K.; Hamada, M. PBSIM: PacBio reads simulator—Toward accurate genome assembly. Bioinformatics 2013, 29, 119–121. [Google Scholar] [CrossRef] [PubMed]
- Mirebrahim, H.; Close, T.J.; Lonardi, S. De novo meta-assembly of ultra-deep sequencing data. Bioinformatics 2015, 31, i9–i16. [Google Scholar] [CrossRef] [PubMed]
- Lonardi, S.; Mirebrahim, H.; Wanamaker, S.; Alpert, M.; Ciardo, G.; Duma, D.; Close, T.J. When less is more: “Slicing” sequencing data improves read decoding accuracy and de novo assembly quality. Bioinformatics 2015, 31, 2972–2980. [Google Scholar] [CrossRef] [PubMed]
- Reen, F.J.; Gutiérrez-Barranquero, J.A.; Dobson, A.D.W.; Adams, C.; O’Gara, F. Emerging concepts promising new horizons for marine biodiscovery and synthetic biology. Mar. Drugs 2015, 13, 2924–2954. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, J.; Flemer, B.; Jackson, S.A.; Lejon, D.P.H.; Morrissey, J.P.; O’Gara, F.; Dobson, A.D.W. Marine metagenomics: New tools for the study and exploitation of marine microbial metabolism. Mar. Drugs 2010, 8, 608–628. [Google Scholar] [CrossRef] [PubMed]
- Wilson, M.C.; Piel, J. Metagenomic approaches for exploiting uncultivated bacteria as a resource for novel biosynthetic enzymology. Chem. Biol. 2013, 20, 636–647. [Google Scholar] [CrossRef] [PubMed]
- Suenaga, H. Targeted metagenomics: A high-resolution metagenomics approach for specific gene clusters in complex microbial communities. Environ. Microbiol. 2012, 14, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Wilson, M.C.; Mori, T.; Rückert, C.; Uria, A.R.; Helf, M.J.; Takada, K.; Gernert, C.; Steffens, U.A.E.; Heycke, N.; Schmitt, S.; et al. An environmental bacterial taxon with a large and distinct metabolic repertoire. Nature 2014, 506, 58–62. [Google Scholar] [CrossRef] [PubMed]
- Baker, M. De novo genome assembly: What every biologist should know. Nat. Methods 2012, 9, 333. [Google Scholar] [CrossRef]
- Sczyrba, A.; Hofmann, P.; Belmann, P.; Koslicki, D. Critical Assessment of Metagenome Interpretation—A benchmark of computational metagenomics software. bioRxiv 2017. [Google Scholar] [CrossRef]
- Medema, M.H.; Kottmann, R.; Yilmaz, P.; Cummings, M.; Biggins, J.B.; Blin, K.; de Bruijn, I.; Chooi, Y.H.; Claesen, J.; Coates, R.C.; et al. Minimum Information about a Biosynthetic Gene cluster. Nat. Chem. Biol. 2015, 11, 625–631. [Google Scholar] [CrossRef] [PubMed]
- Garza, D.R.; Dutilh, B.E. From cultured to uncultured genome sequences: Metagenomics and modeling microbial ecosystems. Cell. Mol. Life Sci. 2015, 72, 4287–4308. [Google Scholar] [CrossRef] [PubMed]
- Klitgord, N.; Segrè, D. Environments that induce synthetic microbial ecosystems. PLoS Comput. Biol. 2010, 6, e1001002. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miller, I.J.; Chevrette, M.G.; Kwan, J.C. Interpreting Microbial Biosynthesis in the Genomic Age: Biological and Practical Considerations. Mar. Drugs 2017, 15, 165. https://doi.org/10.3390/md15060165
Miller IJ, Chevrette MG, Kwan JC. Interpreting Microbial Biosynthesis in the Genomic Age: Biological and Practical Considerations. Marine Drugs. 2017; 15(6):165. https://doi.org/10.3390/md15060165
Chicago/Turabian StyleMiller, Ian J., Marc G. Chevrette, and Jason C. Kwan. 2017. "Interpreting Microbial Biosynthesis in the Genomic Age: Biological and Practical Considerations" Marine Drugs 15, no. 6: 165. https://doi.org/10.3390/md15060165





