Cembrene Diterpenoids with Ether Linkages from Sarcophyton ehrenbergi: An Anti-Proliferation and Molecular-Docking Assessment
Abstract
:1. Introduction
2. Results and Discussion
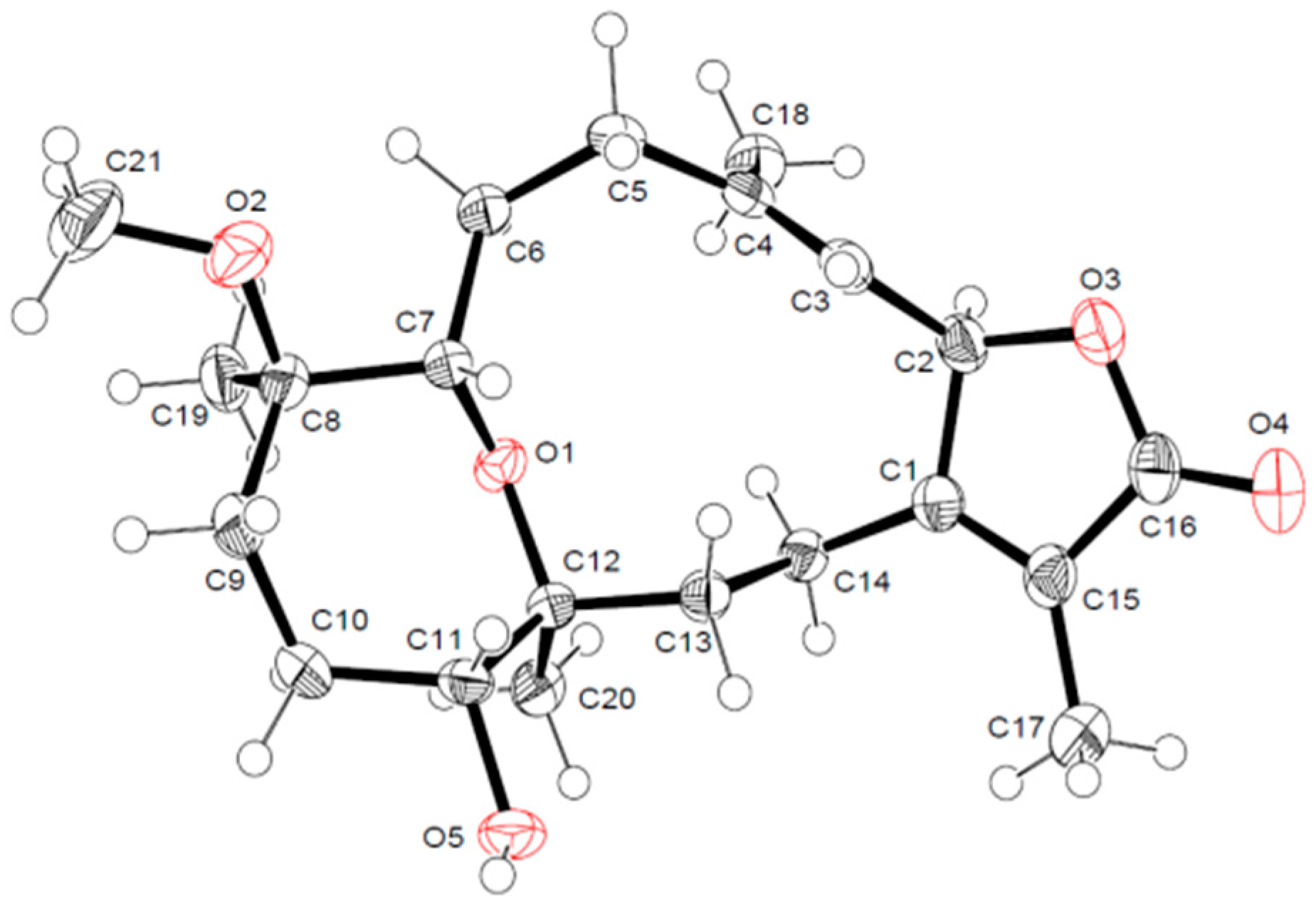
2.1. Identification and Structure Elucidation
2.2. Anti-Proliferative Activity against Cancer Lines
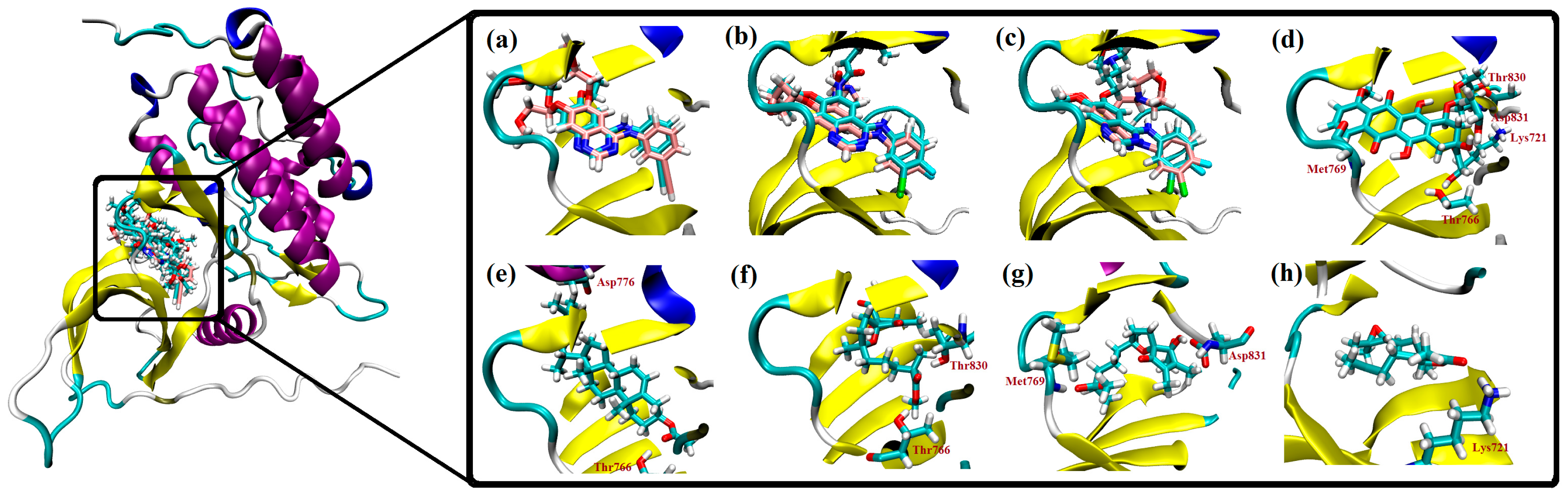
2.3. Molecular Docking Studies
3. Experimental Section
3.1. General Experimental Procedures
3.2. Animal Material
3.3. Extraction and Separation
X-ray Crystallography Data
3.4. Cell Culture
3.5. Cell Proliferation Assay
3.6. Anti-Proliferation Quantitative Analysis
3.7. Computational Methodology
3.7.1. Density Functional Theory Calculations
3.7.2. Molecular Docking Studies
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Yang, B.; Zhou, X.-F.; Lin, X.-P.; Liu, J.; Peng, Y.; Yang, X.-W.; Liu, Y. Cembrane diterpenes chemistry and biological properties. Curr. Org. Chem. 2012, 16, 1512–1539. [Google Scholar] [CrossRef]
- Kobayashi, M.; Nakagawa, T.; Mitsuhashi, H. Marine terpenes and terpenoids. I. Structures of four cembrane-type diterpenes: Sarcophytol-A, sarcophytol-A acetate sarcophytol-B, and sarcophytonin-A, from the soft coral, Sarcophyton glaucum. Chem. Pharm. Bull. 1979, 27, 2382–2387. [Google Scholar] [CrossRef]
- Yang, B.; Liu, J.; Wang, J.; Liao, S.; Liu, Y. Handbook of anticancer drugs from marine origin. In Cytotoxic Cembrane Diterpenoids; Kim, S.-K., Ed.; Springer International Publishing: Cham, Switzerland, 2015. [Google Scholar]
- Abou El-Ezz, R.; Ahmed, S.; Radwan, M.; Ayoub, N.; Afifi, M.; Ross, S.; Szymanski, P.; Fahmy, H.; Khalifa, S. Bioactive cembranoids from the Red Sea soft coral Sarcophyton glaucum. Tetrahedron Lett. 2013, 54, 989–992. [Google Scholar] [CrossRef]
- Liang, L.; Guo, Y. Terpenes from the soft corals of the genus Sarcophyton: Chemistry and biological activities. Chem. Biodivers. 2013, 10, 2161–2196. [Google Scholar] [CrossRef] [PubMed]
- Hegazy, M.E.; El-Beih, A.A.; Moustafa, A.Y.; Hamdy, A.A.; Alhammady, M.A.; Selim, R.M.; Abdel-Rehim, M.; Paré, P.W. Cytotoxic cembranoids from the Red Sea soft coral Sarcophyton glaucum. Nat. Prod. Commun. 2011, 6, 1809–1812. [Google Scholar] [PubMed]
- Hegazy, M.E.; Mohamed, T.A.; Abdel-Latif, F.A.; Alsaid, M.; Shahat, A.A.; Paré, P.W. Trochelioid A and B, new cembranoid diterpenes from the Red Sea soft coral Sarcophyton trocheliophorum. Phytochem. Lett. 2013, 6, 383–386. [Google Scholar] [CrossRef]
- Elkhateeb, A.; El-Beih, A.; Gamal-Eldeen, A.; Alhammady, M.; Ohta, S.; Paré, P.; Hegazy, M.E. New terpenes from the Egyptian soft coral Sarcophyton ehrenbergi. Mar. Drugs 2014, 12, 1977–1986. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.-H.; Li, Z.-Y.; Guo, Y.-W. Further new cembranoid diterpenes from the Hainan soft coral Sarcophyton latum. Helv. Chim. Acta 2007, 90, 1574. [Google Scholar] [CrossRef]
- Yan, X.-H.; Feng, L.-Y.; Guo, Y.-W. Further new cembrane diterpenes from the Hainan soft coral Sarcophyton latum. Chin. J. Chem. 2008, 26, 150–152. [Google Scholar] [CrossRef]
- Rodriguez, A.D.; Shi, J.G. The first cembrane-pseudopterane cycloisomerization. J. Org. Chem. 1998, 63, 420–421. [Google Scholar] [CrossRef] [PubMed]
- Qin, S.; Huang, H.; Guo, Y.W. A new cembranoid from the Hainan soft coral Sinularia sp. J. Asian Nat. Prod. Res. 2008, 10, 1075–1079. [Google Scholar] [CrossRef] [PubMed]
- Hegazy, M.E.F.; Mohamed, T.A.; Alhammady, M.A.; Shaheen, A.M.; Reda, E.H.; Elshamy, A.I.; Aziz, M.; Paré, P.W. Molecular architecture and biomedical leads of terpenes from Red Sea marine invertebrate. Mar. Drugs 2015, 13, 3154–3181. [Google Scholar] [CrossRef] [PubMed]
- Torre, L.A.; Bray, F.; Siegel, R.L.; Ferlay, J.; Lortet-Tieulent, J.; Jemal, A. Global cancer statistics, 2012. CA Cancer J. Clin. 2015, 65, 87–108. [Google Scholar] [CrossRef] [PubMed]
- Society, A.C. Cancer Facts & Figures. 2016. Available online: http://www.cancer.org/acs/groups/content/@research/documents/document/acspc-047079.pdf (accessed on 15 June 2017).
- Hegazy, M.E.F.; Mohamed, T.A.; Elshamy, A.I.; Alhammady, M.A.; Ohta, S.; Paré, P. Casbane Diterpenes from Red Sea Coral Sinularia polydactyla. Molecules 2016, 21, 308. [Google Scholar] [CrossRef] [PubMed]
- Parsons, S.; Flack, H.D.; Wagner, T. Use of intensity quotients and differences in absolute structure refinement. Acta Cryst. 2013, 69, 249–259. [Google Scholar] [CrossRef] [PubMed]
- Parsons, S.; Flack, H. Precise absolute-structure determination in light-atom crystals. Acta Cryst. 2004, 60, s61. [Google Scholar] [CrossRef]
- Sawant, S.; Sylvester, P.; Avery, M.; Desai, P.; Youssef, D.; El Sayed, K. Bioactive rearranged and halogenated semisynthetic derivatives of the marine natural product sarcophine. J. Nat. Prod. 2004, 67, 2017–2023. [Google Scholar] [CrossRef] [PubMed]
- Xi, Z.; Bie, W.; Chen, W.; Liu, D.; Ofwegen, L.V.; Proksch, P.; Lin, W. Sarcophyolides B–E, new cembranoids from the soft coral Sarcophyton elegans. Mar. Drugs 2013, 11, 3186–3196. [Google Scholar] [CrossRef] [PubMed]
- Su, J.-H.; Lu, Y.; Lin, W.-Y.; Wang, W.-H.; Sung, P.-J.; Sheu, J.-H. A cembranoid, trocheliophorol, from the cultured soft coral Sarcophyton trocheliophorum. Chem. Lett. 2010, 39, 172–173. [Google Scholar] [CrossRef]
- Yang, B.; Wei, X.; Huang, J.; Lin, X.; Liu, J.; Liao, S.; Wang, J.; Zhou, X.; Wang, L.; Liu, Y. Sinulolides A–H, new cyclopentenone and butenolide derivatives from soft coral Sinularia sp. Mar. Drugs 2014, 12, 5316–5327. [Google Scholar] [CrossRef] [PubMed]
- Su, J.; Yang, R.; Zeng, L. Sardisterol, a new polyhydroxylated sterol from the soft coral Sarcophyton digitatum Moser. Chin. J. Chem. 2001, 19, 515–517. [Google Scholar] [CrossRef]
- Dutta, P.R.; Maity, A. Cellular responses to EGFR inhibitors and their relevance to cancer therapy. Cancer Lett. 2007, 254, 165–177. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Wang, X.-Y.; Lv, P.-C.; Zhu, H.-L. Discovery of a series of novel phenylpiperazine derivatives as EGFR TK inhibitors. Sci. Rep. 2015, 5, 13934. [Google Scholar] [CrossRef] [PubMed]
- Sheldrick, G.M. SHELXS-97: Program for Crystal Structure Solution; University of Göttingen: Göttingen, Germany, 1997. [Google Scholar]
- Sheldrick, G.M. SHELXL-2013: Program for Crystal Structure Refinement; University of Göttingen: Göttingen, Germany, 2013. [Google Scholar]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
- Dutta, A.; Bandyopadhyay, S.; Mandal, C.; Chatterjee, M. Development of a modified MTT assay for screening antimonial resistant field isolates of Indian visceral leishmaniasis. Parasitol. Int. 2005, 54, 119–122. [Google Scholar] [CrossRef] [PubMed]
- Hawkins, P.C.D.; Skillman, A.G.; Warren, G.L.; Ellingson, B.A.; Stahl, M.T. Conformer Generation with OMEGA: Algorithm and Validation Using High Quality Structures from the Protein Databank and Cambridge Structural Database. J. Chem. Inf. Model. 2010, 50, 572. [Google Scholar] [CrossRef] [PubMed]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. Gaussian09; Revision E.01; Gaussian Inc.: Wallingford, CT, USA, 2009. [Google Scholar]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed]
- Stamos, J.; Sliwkowski, M.X.; Eigenbrot, C. Structure of the Epidermal Growth Factor Receptor Kinase Domain Alone and in Complex with a 4-Anilinoquinazoline Inhibitor. J. Biol. Chem. 2002, 277, 46265–46272. [Google Scholar] [CrossRef] [PubMed]
- Forli, S.; Huey, R.; Pique, M.E.; Sanner, M.F.; Goodsell, D.S.; Olson, A.J. Computational protein-ligand docking and virtual drug screening with the AutoDock suite. Nat. Protoc. 2016, 11, 905–919. [Google Scholar] [CrossRef] [PubMed]
- SZYBKI, version 1.9.0.3; OpenEye Scientific Software: Santa Fe, NM, USA, 2016.
- Case, D.; Babin, V.; Berryman, J.; Betz, R.; Cai, Q.; Cerutti, D.; Cheatham, T.; Darden, T.; Duke, R.; Gohlke, H.; et al. Amber 14; University of California: San Francisco, CA, USA, 2014. [Google Scholar]
- Wang, J.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and testing of a general amber force field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef] [PubMed]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the Accuracy of Protein Side Chain. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef] [PubMed]


 ) correlations of 1–3.
) correlations of 1–3.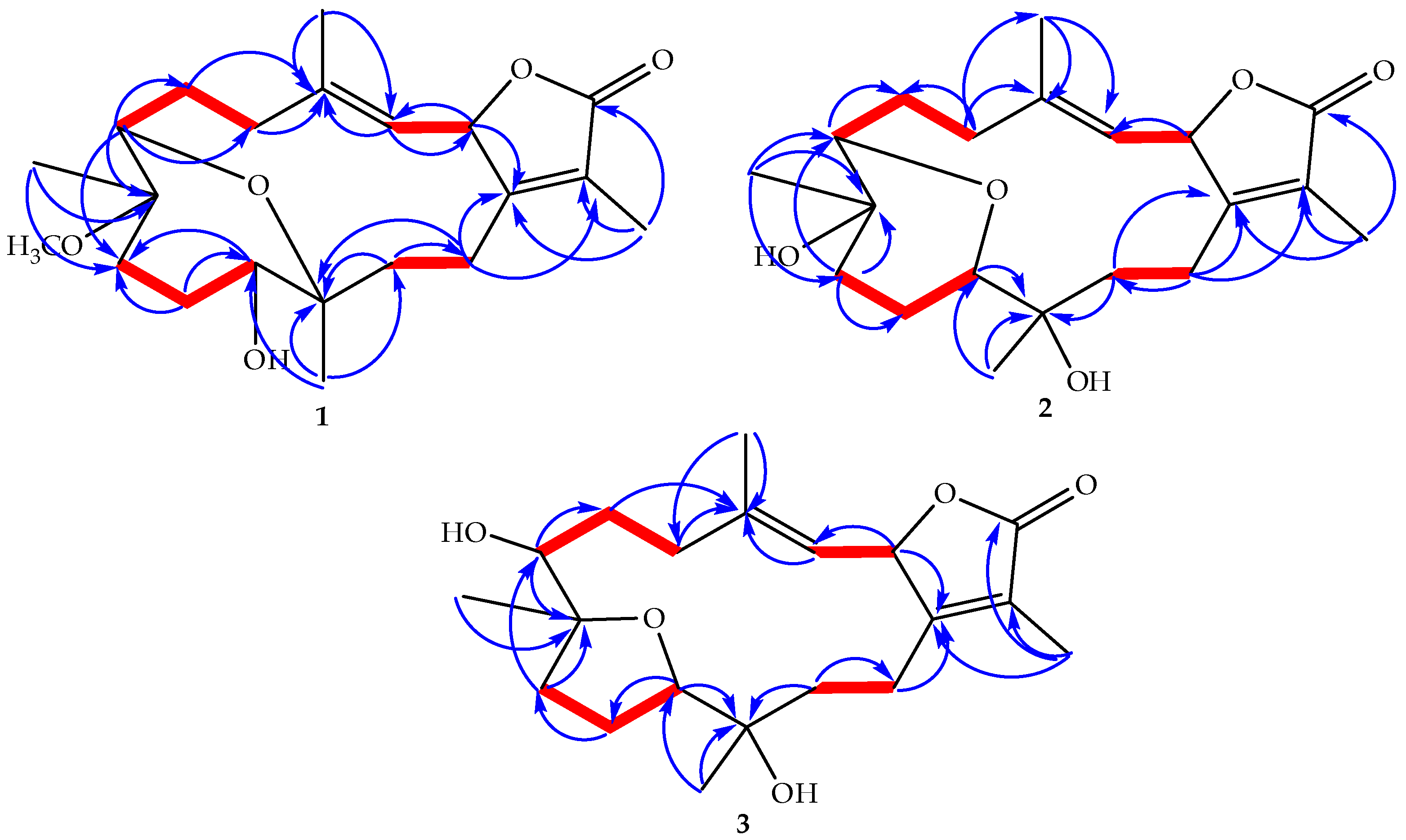
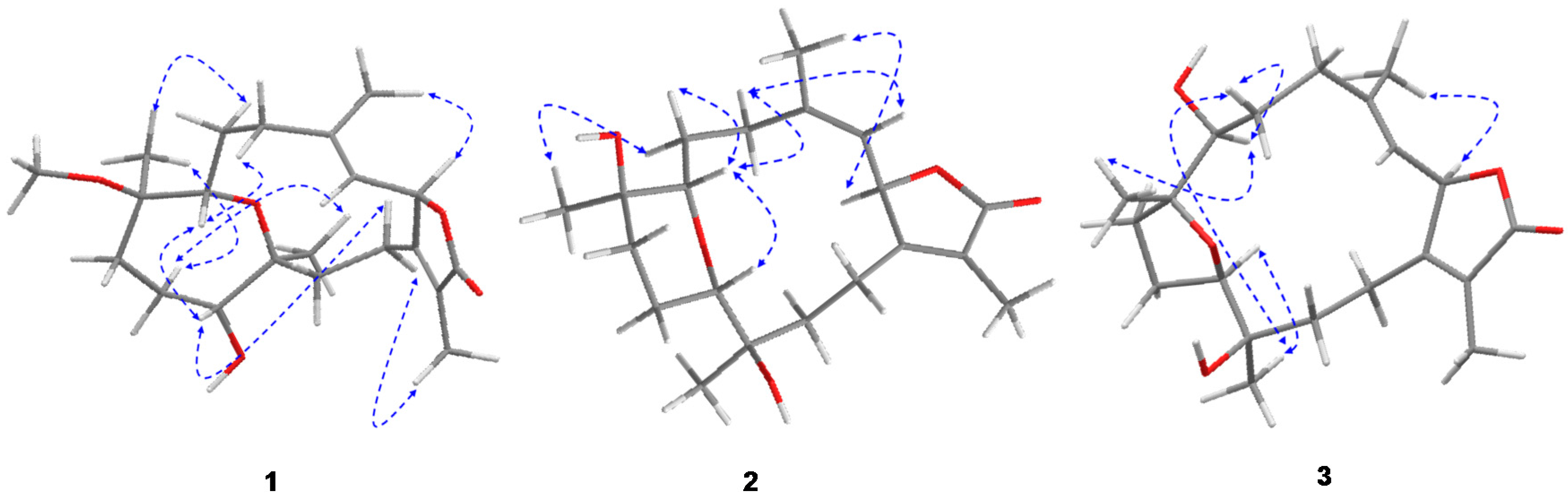

| No. | 1 | 2 | 3 | |||
|---|---|---|---|---|---|---|
| δH | δC | δH | δC | δH | δC | |
| 1 | ------ | 164.3 | ------ | 164.6 | ------ | 164.4 |
| 2 | 5.50 d (10.1) | 81.0 | 5.49 dd (10.1; 1.8) | 81.7 | 5.51 br d (10.3) | 79.6 |
| 3 | 4.94 d (10.1) | 119.4 | 4.99 d (10.1) | 120.0 | 4.87 br d (10.3) | 117.8 |
| 4 | ------ | 141.6 | ------ | 142.6 | ------ | 146.0 |
| 5 | 2.07 t (13.1) 2.30 m | 40.9 | 2.17 br t (13.1) 2.47 dd (10.1; 13.1) | 38.8 | 2.09 br t (13.0) 2.41 br d (13.0) | 40.3 |
| 6 | 2.14 dd (6.8), 1.44 m | 27.6 | 1.66 m; 2.20 m | 22.9 | 1.88 m; 1.58 m | 24.9 |
| 7 | 3.24 d (6.9) | 78.5 | 3.44 dd (11.6; 3.6) | 69.7 | 3.01 br d (10.0) | 86.8 |
| 8 | ------ | 80.0 | ------ | 73.6 | ------ | 69.4 |
| 9 | 1.53 m; 1.91 m | 36.4 | 1.60 m; 2.40 m | 36.6 | 1.88 m; 2.41 br d (13.0) | 39.9 |
| 10 | 1.79 m; 2.15 m | 28.5 | 1.61 m; 1.90 m | 19.9 | 1.71 br d (10.9) 1.51 br t (10.9) | 23.1 |
| 11 | 3.37 m | 79.0 | 3.37 br d (11.9) | 85.1 | 3.30 br d (10.9) | 78.5 |
| 12 | ------ | 78.3 | ------ | 70.1 | ------ | 72.8 |
| 13 | 1.45 m; 2.37 m | 34.6 | 1.59 m 1.78 dd (12.5; 3.7) | 31.2 | 1.41 td (13.0; 5.8) 1.78 br t (13.0; 2.0) | 35.0 |
| 14 | 1.95 br t (12.2) 2.31 td (12.2, 7.0) | 20.8 | 2.00 br t (12.8) 2.58 td (12.8; 7.0) | 21.1 | 2.17 br t (13.0); 2.67 m | 21.1 |
| 15 | ------ | 122.3 | ------ | 122.3 | ------ | 122.5 |
| 16 | ------ | 175.9 | ------ | 176.0 | ------ | 175.6 |
| 17 | 1.76 s | 8.8 | 1.83 s | 8.7 | 1.82 s | 8.7 |
| 18 | 1.86 s | 17.0 | 1.92 br s | 16.7 | 1.85 br s | 16.1 |
| 19 | 1.05 s | 17.1 | 1.22 s | 22.1 | 1.15 s | 20.0 |
| 20 | 0.98 s | 17.6 | 1.13 s | 25.3 | 1.16 s | 23.8 |
| 21 | 3.13 s | 49.0 | ||||
| Compound | A549 IC50(µM) | Caco-2 IC50(µM) | HepG2 IC50(µM) |
|---|---|---|---|
| 1 | 50.1 | >100 | 98.6 |
| 2 | 76.4 | >100 | >100 |
| 3 | 50.8 | >100 | 53.8 |
| 4 | 91.5 | >100 | >100 |
| 5 | 62.2 | >100 | 79.3 |
| 6 | 37.0 | 79.2 | 70.2 |
| 7 | 43.6 | 99.2 | 63.1 |
| 8 | 27.3 | >100 | 56.8 |
| Doxorubicin HCl | 0.62 | 1.40 | 2.10 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hegazy, M.-E.F.; Elshamy, A.I.; Mohamed, T.A.; Hamed, A.R.; Ibrahim, M.A.A.; Ohta, S.; Paré, P.W. Cembrene Diterpenoids with Ether Linkages from Sarcophyton ehrenbergi: An Anti-Proliferation and Molecular-Docking Assessment. Mar. Drugs 2017, 15, 192. https://doi.org/10.3390/md15060192
Hegazy M-EF, Elshamy AI, Mohamed TA, Hamed AR, Ibrahim MAA, Ohta S, Paré PW. Cembrene Diterpenoids with Ether Linkages from Sarcophyton ehrenbergi: An Anti-Proliferation and Molecular-Docking Assessment. Marine Drugs. 2017; 15(6):192. https://doi.org/10.3390/md15060192
Chicago/Turabian StyleHegazy, Mohamed-Elamir F., Abdelsamed I. Elshamy, Tarik A. Mohamed, Ahmed R. Hamed, Mahmoud A. A. Ibrahim, Shinji Ohta, and Paul W. Paré. 2017. "Cembrene Diterpenoids with Ether Linkages from Sarcophyton ehrenbergi: An Anti-Proliferation and Molecular-Docking Assessment" Marine Drugs 15, no. 6: 192. https://doi.org/10.3390/md15060192







