Effect of Pentacyclic Guanidine Alkaloids from the Sponge Monanchora pulchra on Activity of α-Glycosidases from Marine Bacteria
Abstract
:1. Introduction
2. Results and Discussion
2.1. Identification of the Compounds
2.2. Effect of Monanchomycalin B, Monanchocidin A, and Normonanchocidin A on Activity of Two Glycosidases
E + nI ⇌ [E In] → E‒In,
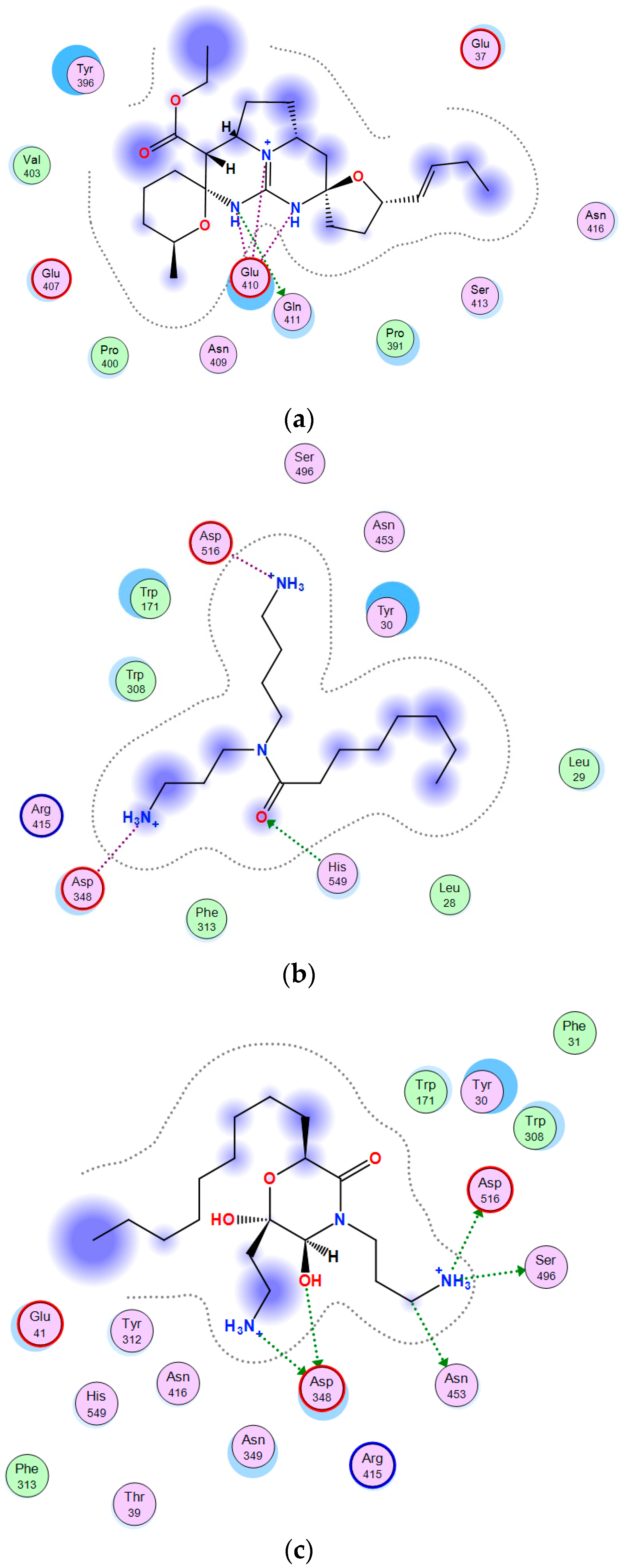
2.3. Theoretical Model of the Guanidine Alkaloids Complexes with α-Galactosidase
3. Materials and Methods
3.1. Materials
3.2. Experimental Equipment
3.3. Collection and Identification of Sponge Material
3.4. Isolation and Purification of Compounds
3.5. Production of Recombinant Enzymes
3.5.1. Production and Purification of Recombinant α-d-galactosidase
3.5.2. Production and Purification of Recombinant α-Nacetylgalactosaminidase
3.5.3. Enzyme and Protein Assays
3.6. Effects of Pentacyclic Guanidine Alkaloids on Glycosidases of Marine Bacteria
3.6.1. The Effects of Monanchomycalin B, Monanchocidin A, and Normonanchocidin A on Glyctosidases
3.6.2. The Irreversibility of Monanchomycalin B Inhibition
3.6.3. The Assay of α-PsGal Inhibition by Monanchomycalin B, Monanchocidin A, and Normonanchocidin A
3.6.4. The Kinetic Parameters of Inactivation
3.6.5. Protection of α-PsGal Inactivation by d-galactose
3.7. Theoretical Models of α-PsGal Complexes with Guanidine Alkaloids
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Asano, N. Glycosidase inhibitors: Update and perspectives on practical use. Glycobiology 2003, 13, 93R–104R. [Google Scholar] [CrossRef] [PubMed]
- Skropeta, D.; Pastro, N.; Zivanovic, A. Kinase inhibitors from marine sponges. Mar. Drugs 2011, 9, 2131–2154. [Google Scholar] [CrossRef] [PubMed]
- Ruocco, N.; Costantini, S.; Palumbo, F.; Constantini, M. Marine sponges and bacteria as challenging sources of enzyme inhibitors for pharmacological applications. Mar. Drugs 2017, 15, 173. [Google Scholar] [CrossRef] [PubMed]
- Bakunina, I.Y.; Balabanova, L.A.; Pennacchio, A.; Trincone, A. Hooked on α-d-galactosidases: From biomedicine to enzymatic synthesis. Crit. Rev. Biotechnol. 2016, 36, 233–245. [Google Scholar] [CrossRef] [PubMed]
- Bakunina, I.Y.; Ivanova, E.P.; Nedashkovskaya, O.I.; Gorshkova, N.M.; Elyakova, L.A.; Mikhailov, V.V. Search for α-d-galactosidase producers among marine bacteria of the genus Alteromonas. Prikl. Biokh. Mikrobiol. (Moscow) 1996, 32, 624–628. [Google Scholar]
- Ivanova, E.P.; Bakunina, I.Y.; Nedashkovskaya, O.I.; Gorshkova, N.; Mikhailov, V.V.; Elyakova, L.A. Incidence of marine microorganisms producing β-N-acetylglucosaminidases, α-d-galactosidases and α-N-acetylgalactosaminidases. Rus. J. Mar. Biol. 1998, 24, 365–372. [Google Scholar]
- Bakunina, I.Y.; Nedashkovskaya, O.I.; Alekseeva, S.A.; Ivanova, E.P.; Romanenko, L.A.; Gorshkova, N.M.; Isakov, V.V.; Zvyagintseva, T.N.; Mikhailov, V.V. Degradation of fucoidan by the marine proteobacterium Pseudoalteromonas citrea. Microbiology (Moscow) 2002, 71, 41–47. [Google Scholar] [CrossRef]
- Bakunina, I.Y.; Nedashkovskaya, O.I.; Kim, S.B.; Zvyagintseva, T.N.; Mihailov, V.V. Diversity of glycosidase activities in the bacteria of the phylum Bacteroidetes isolated from marine algae. Microbiology (Moscow) 2012, 81, 688–695. [Google Scholar] [CrossRef]
- Bakunina, I.Y.; Sova, V.V.; Nedashkovskaya, O.I.; Kuhlmann, R.A.; Likhosherstov, L.M.; Martynova, M.I.; Mihailov, V.V.; Elyakova, L.A. α-d-galactosidase of the marine bacterium Pseudoalteromonas sp. KMM 701. Biochemisrty (Moscow) 1998, 63, 1209–1215. [Google Scholar]
- Balabanova, L.A.; Bakunina, I.Y.; Nedashkovskaya, O.I.; Makarenkova, I.D.; Zaporozhets, T.S.; Besednova, N.N.; Zvyagintseva, T.N.; Rasskazov, V.A. Molecular characterization and therapeutic potential of a marine bacterium Pseudoalteromonas sp. KMM 701 α-d-galactosidase. Mar. Biotechnol. 2010, 12, 111–120. [Google Scholar] [CrossRef]
- Terentieva, N.A.; Timchenko, N.F.; Balabanova, L.A.; Golotin, V.A.; Belik, A.A.; Bakunina, I.Y.; Didenko, L.V.; Rasskazov, V.A. The influence of enzymes on the formation of bacterial biofilms. Health Med. Ecol. Sci. 2015, 60, 86–93. [Google Scholar]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The Carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, D490–D495. [Google Scholar] [CrossRef] [PubMed]
- Bakunina, I.Y.; Nedashkovskaya, O.I.; Kim, S.B.; Zvyagintseva, T.N.; Mikhailov, V.V. Distribution of α-N-acetylgalactosaminidases among marine bacteria of the phylum Bacteroidetes, epiphytes of marine algae of the Seas of Okhotsk and Japan. Microbiology (Moscow) 2012, 81, 375–380. [Google Scholar] [CrossRef]
- Bakunina, I.Y.; Nedashkovskaya, O.I.; Balabanova, L.A.; Zvyagintseva, T.N.; Rasskasov, V.A.; Mikhailov, V.V. Comparative analysis of glycoside hydrolases activities from phylogenetically diverse marine bacteria of the genus Arenibacter. Mar. Drugs 2013, 11, 1977–1998. [Google Scholar] [CrossRef] [PubMed]
- Bakunina, I.Y.; Kuhlmann, R.A.; Likhosherstov, L.M.; Martynova, M.D.; Nedashkovskaya, O.I.; Mikhailov, V.V.; Elyakova, L.A. α-N-Acetylgalactosaminidase from marine bacterium Arenibacter latericius KMM 426T removing blood type specificity of A erythrocytes. Biochemistry (Moscow) 2002, 67, 689–695. [Google Scholar] [CrossRef]
- Blunt, J.W.; Carroll, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2018, 35, 8–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Li, X.-W.; Guo, Y.-W. Recent advances in the isolation, synthesis and biological activity of marine guanidine alkaloids. Mar. Drugs 2017, 15, 324. [Google Scholar] [CrossRef]
- Sfecci, E.; Lacour, T.; Amade, P.; Mehiri, M. Polycyclic guanidine alkaloids from Poecilosclerida Marine Sponges. Mar. Drugs 2016, 14, 77. [Google Scholar] [CrossRef]
- Shrestha, S.; Sorolla, A.; Fromont, J.; Blancafort, P.; Flematti, G.R. Crambescidin 800, isolated from the marine sponge Monanchora viridis, induces cell cycle arrest and apoptosis in triple-negative breast cancer cells. Mar. Drugs 2018, 16, 53. [Google Scholar] [CrossRef]
- Guzii, A.G.; Makarieva, T.N.; Denisenko, V.A.; Dmitrenok, P.S.; Kuzmich, A.S.; Dyshlovoy, S.A.; Krasokhin, V.B.; Stonik, V.A. Monanchocidin: A new apoptosis-inducing polycyclic guanidine alkaloid from the marine sponge Monanchora pulchra. Org. Lett. 2010, 12, 4292–4295. [Google Scholar] [CrossRef]
- Makarieva, T.N.; Tabakmaher, K.M.; Guzii, A.G.; Denisenko, V.; Dmitrenok, P.S.; Kuzmich, A.S.; Lee, H.-S.; Stonik, V.A. Monanchomycalins A and B, unusual guanidine alkaloids from the sponge Monanchora pulchra. Tetrahedron Lett. 2012, 53, 4228–4231. [Google Scholar] [CrossRef]
- Tabakmakher, K.M.; Makarieva, T.N.; Denisenko, V.A.; Guzii, A.G.; Dmitrenok, P.S.; Kuzmich, A.S.; Stonik, V.A. Normonanchocidins A, B and D, new pentacyclic guanidine glkaloids from the Far-Eastern marine sponge Monanchora pulchra. Nat. Prod. Commun. 2015, 10, 913–916. [Google Scholar] [PubMed]
- Dubrovskaya, Y.V.; Makarieva, T.N.; Shubina, L.K.; Bakunina, I.Y. Effect of pentacyclic guanidine alkaloids from the marine sponge Monanchora pulchra Lambe, 1894 on activity of natural β-1,3-d-glucanase from the marine fungus Chaetomium indicum Corda, 1840 and the marine bivalve mollusk Spisula sachalinensis, Schrenck, 1861. Rus. J. Mar. Biol. 2018, 44, 127–134. [Google Scholar]
- Bakunina, I.; Slepchenko, L.; Anastyuk, S.; Isakov, V.; Likhatskaya, G.; Kim, N.; Tekutyeva, L.; Son, O.; Balabanova, L. Characterization of properties and transglycosylation abilities of recombinant α-galactosidase from cold adapted marine bacterium Pseudoalteromonas KMM 701 and its С494N, D451A mutants. Mar. Drugs 2018, 16, 349. [Google Scholar] [CrossRef] [PubMed]
- Parsons, Z.D.; Gates, K.S. Redox regulation of protein tyrosine phosphatases: Methods for kinetic analysis of covalent enzyme inactivation. Methods Enzymol. 2013, 528, 129–154. [Google Scholar] [PubMed]
- Morrison, J.F. The slow-binding and slow, tight-binding inhibition of enzyme-catalyzed reactions. Trends Biochem. Sci. 1982, 7, 102–105. [Google Scholar] [CrossRef]
- Powers, J.C.; Asgian, J.L.; Ekici, O.D.; James, K.E. Irreversible inhibitors of serine, cysteine, and threonine proteases. Chem. Rev. 2002, 102, 4639–4750. [Google Scholar] [CrossRef]
- Esteban, G.; Allan, J.; Samadi, A.; Mattevi, A.; Unzeta, M.; Marco-Contelles, J.; Binda, C.; Ramsay, R.R. Kinetic and structural analysis of the irreversible inhibition of human monoamine oxidases by ASS234, a multi-target compound designed for use in Alzheimer’s disease. Biochim. Biophys. Acta 2014, 1844, 1104–1110. [Google Scholar] [CrossRef]
- Ramsay, R.R.; Tipton, K.F. Assessment of enzyme inhibition: A review with examples from the development of monoamine oxidase and cholinesterase inhibitory drugs. Molecules 2017, 22, 1192. [Google Scholar] [CrossRef]
- Dyshlovoy, S.A.; Hauschild, J.; Amann, K.; Tabakmakher, K.M.; Venz, S.; Walther, R.; Guzii, A.G.; Makarieva, T.N.; Shubina, L.K.; Fedorov, S.N.; et al. Marine alkaloid monanchocidin A overcomes drug resistance by induction of autophagy and lysosomal membrane permeabilization. Oncotarget 2015, 6, 17328–17341. [Google Scholar] [CrossRef]
- Bakunina, I.Y.; Kol’tsova, E.A.; Pokhilo, N.D.; Shestak, O.P.; Yakubovskaya, A.Y.; Zvyagintseva, T.N.; Anufriev, V.F. Effect of 5-hydroxy- and 5,8-dihydroxy-1,4-naphthoquinones on the hydrolytic activity of alpha-galactosidase. Chem. Nat. Comp. 2009, 45, 69–73. [Google Scholar] [CrossRef]
- Gote, M.M.; Khan, M.I.; Gokhale, D.V.; Bastawde, K.B.; Khire, J.M. Purification, characterization and substrate specificity of thermostable α-galactosidase from Bacillus stearothermophilus (NCIM-5146). Process Biochem. 2006, 41, 1311–1317. [Google Scholar] [CrossRef]
- Borisova, A.S.; Reddy, S.K.; Ivanen, D.R.; Bobrov, K.S.; Eneyskaya, E.V.; Rychkov, G.N.; Sandgren, M.; Stålbrand, H.; Sinnott, M.L.; Kulminskaya, A.A.; et al. The method of integrated kinetics and its applicability to the exo-glycosidase-catalyzed hydrolysis of p-nitrophenyl glycosides. Carbohydr. Res. 2015, 412, 43–49. [Google Scholar] [CrossRef] [PubMed]
- MOE, 2018.01; Chemical Computing Group ULC: 1010 Sherbrooke St. West, Suite #910, Montreal, QC, Canada, 2018.
- Bakunina, I.Y.; Balabanova, L.A.; Golotin, V.A.; Slepchenko, L.A.; Isakov, V.V.; Rasskazov, V.V. Stereochemical course of hydrolytic reaction catalyzed by alpha-galactosidase from cold adaptable marine bacterium of genus Pseudoalteromonas. Front. Chem./Chem. Biol. 2014, 2, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Golotin, V.A.; Balabanova, L.A.; Noskova, Y.A.; Slepchenko, L.A.; Bakunina, I.Y.; Vorobieva, N.S.; Terentieva, N.A.; Rasskazov, V.A. Optimization of cold-adapted α-d-galactosidase expression in Escherichia coli. Protein Expr. Purif. 2016, 123, 14–18. [Google Scholar] [CrossRef] [PubMed]
- Balabanova, L.A.; Golotin, V.А.; Bakunina, I.Y.; Slepchenko, L.V.; Isakov, V.V.; Podvolotskaya, A.B.; Rasskazov, V.А. Recombinant α-N-Acetylgalactosaminidase from marine bacterium modifying A erythrocyte antigens. Acta Nat. 2015, 7, 117–120. [Google Scholar]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Varfolomeev, S.D. Khimicheskaya enzimologiya (Chemical Enzymology); Akademiya: Moscow, Russian, 2005. [Google Scholar]
- Shared Resource Center Far Eastern Computing Resource of Institute of Automation and Control Processes Far Eastern Branch of the Russian Academy of Sciences (IACP FEB RAS). Available online: https://cc.dvo.ru (accessed on 22 June 2016).




| Enzyme | H2O | Monanchomycalin B | Monanchocidin A | Normonanchocidin A |
|---|---|---|---|---|
| α-PsGal | 100 | 0.21 | 2.7 | 1.7 |
| α-NaGa | 100 | 101.5 | 98.5 | 83.0 |
| Monanchomycalin B (μM) | Residual Activity (%) | |
|---|---|---|
| Before Dialysis | After Dialysis | |
| 0 | 100 | 38 |
| 18 | 0 | 0 |
| Inhibitor | kinact (min−1) | Ki (μM) | n |
|---|---|---|---|
| Monanchomycalin B | 0.166 ± 0.029 | 7.70 ±0.62 | 4.64 ± 1.21 |
| Monanchocidin A | 0.08 ± 0.003 | 15.08 ± 1.60 | 2.1 ± 0.47 |
| Normonanchocidin A | 0.026 ± 0.000 | 4.15 ± 0.01 | 4.55 ± 0.02 |
| Concentration (μM) | kobs (min−1) | kobsGal (min–1) 1 |
|---|---|---|
| 11.4 | 0.152 ± 0.005 | 0.082 ± 0.006 |
| 14.2 | 0.178 ± 0.013 | 0.097 ± 0.008 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bakunina, I.; Likhatskaya, G.; Slepchenko, L.; Balabanova, L.; Tekutyeva, L.; Son, O.; Shubina, L.; Makarieva, T. Effect of Pentacyclic Guanidine Alkaloids from the Sponge Monanchora pulchra on Activity of α-Glycosidases from Marine Bacteria. Mar. Drugs 2019, 17, 22. https://doi.org/10.3390/md17010022
Bakunina I, Likhatskaya G, Slepchenko L, Balabanova L, Tekutyeva L, Son O, Shubina L, Makarieva T. Effect of Pentacyclic Guanidine Alkaloids from the Sponge Monanchora pulchra on Activity of α-Glycosidases from Marine Bacteria. Marine Drugs. 2019; 17(1):22. https://doi.org/10.3390/md17010022
Chicago/Turabian StyleBakunina, Irina, Galina Likhatskaya, Lubov Slepchenko, Larissa Balabanova, Liudmila Tekutyeva, Oksana Son, Larisa Shubina, and Tatyana Makarieva. 2019. "Effect of Pentacyclic Guanidine Alkaloids from the Sponge Monanchora pulchra on Activity of α-Glycosidases from Marine Bacteria" Marine Drugs 17, no. 1: 22. https://doi.org/10.3390/md17010022





