Marine Bacteria from Rocas Atoll as a Rich Source of Pharmacologically Active Compounds
Abstract
1. Introduction
2. Results and Discussion
2.1. Bacteria Recovered from Rocas Atoll: Cytotoxicity and Identification
2.2. Chemical Diversity Produced by Bacteria from Rocas Atoll
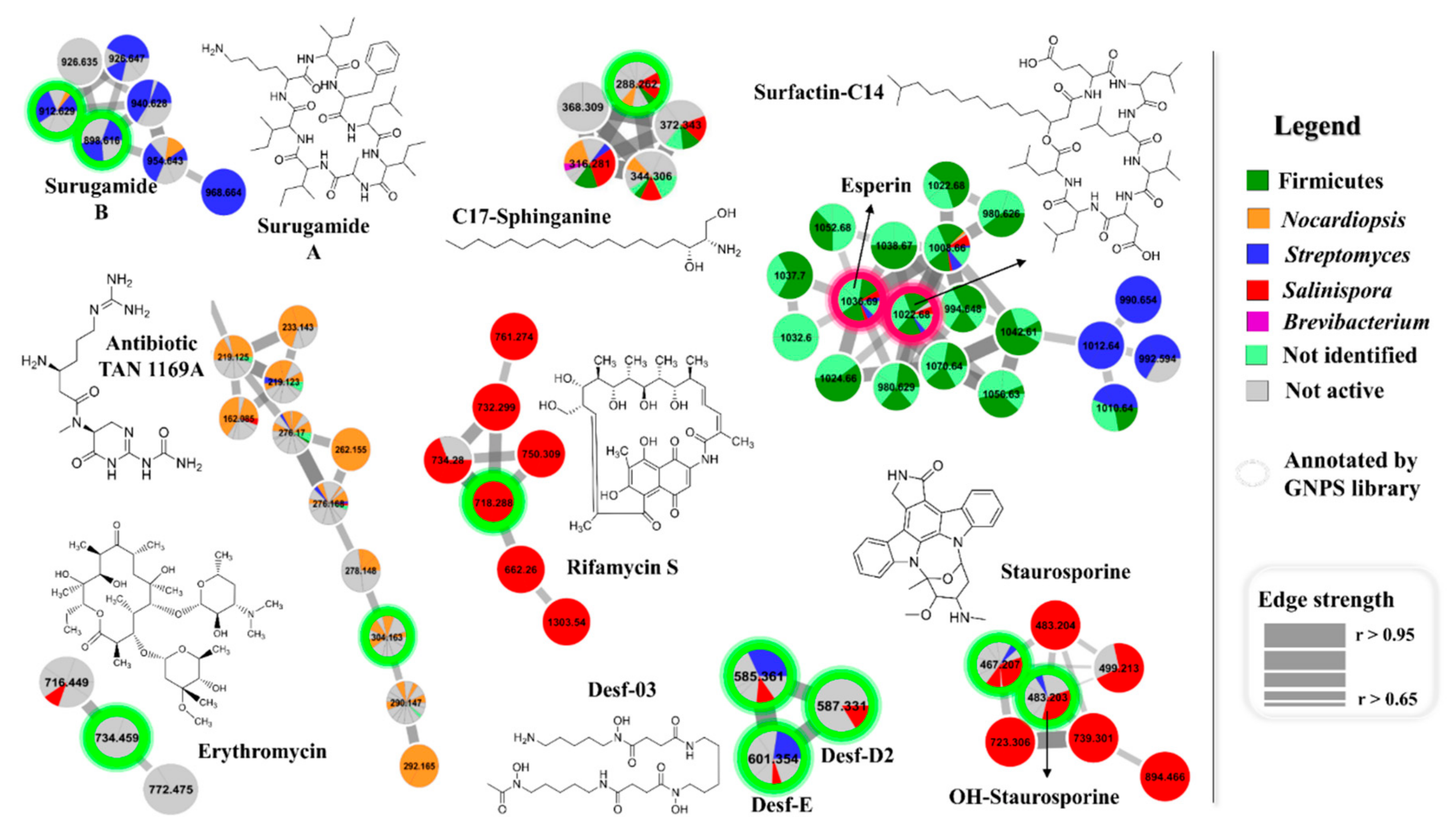
2.2.1. Molecular Networking
2.2.2. Chemometric Analysis
2.3. Molecular Networking vs. Cytotoxicity
2.4. Glycosylated Compounds Produced by BRB-302
3. Materials and Methods
3.1. Sample Collection
3.2. Bacteria Isolation and Cultivation
3.3. Crude Extract Production
3.4. Cultivation and Fractionation of BRB-302 Strain
3.5. Cytotoxic Assay
3.6. DNA Extraction, 16S rRNA Amplification, and Sequencing
3.7. Phylogenetic Analysis
3.8. Metabolomic Fingerprint by HPLC-MS/MS
3.9. Molecular Networking
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Blunt, J.W.; Carroll, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2018, 35, 8–53. [Google Scholar] [CrossRef]
- Soldatou, S.; Baker, B.J. Cold-water marine natural products, 2006 to 2016. Nat. Prod. Rep. 2017, 34, 585–626. [Google Scholar] [CrossRef]
- Miloslavich, P.; Klein, E.; Díaz, J.M.; Hernández, C.E.; Bigatti, G.; Campos, L.; Artigas, F.; Castillo, J.; Penchaszadeh, P.E.; Neill, P.E.; et al. Marine Biodiversity in the Atlantic and Pacific Coasts of South America: Knowledge and Gaps. PLoS ONE 2011, 6, e14631. [Google Scholar] [CrossRef]
- Paiva, S.V.; Oliveira Filho, R.R.D.; Lotufo, T.M. Ascidians from Rocas Atoll, northeast Brazil. Front. Mar. Sci. 2015, 2, 1–20. [Google Scholar] [CrossRef][Green Version]
- Azevedo, F.; Padua, A.; Moraes, F.; Rossi, A.; Muricy, G.; Klautau, M. Taxonomy and phylogeny of calcareous sponges (Porifera: Calcarea: Calcinea) from Brazilian mid-shelf and oceanic islands. Zootaxa 2017, 4311, 301–344. [Google Scholar] [CrossRef]
- Alvarez-Yela, A.C.; Mosquera-Rendón, J.; Noreña, P.A.; Cristancho, M.; López-Alvarez, D. Microbial Diversity Exploration of Marine Hosts at Serrana Bank, a Coral Atoll of the Seaflower Biosphere Reserve. Front. Mar. Sci. 2019, 6, 338. [Google Scholar] [CrossRef]
- Coimbra, J.C.; Carreño, A. Richness and palaeo-zoogeographical significance of the benthic Ostracoda (Crustacea) from the oceanic Island of Trindade and Rocas Atoll, Brazil. Rev. Bras. Paleontolog. 2012, 15, 189–202. [Google Scholar] [CrossRef]
- Netto, S.A.; Attrill, M.J.; Warwick, R.M. The relationship between benthic fauna, carbonate sediments and reef morphology in reef-flat tidal pools of Rocas Atoll (north-east Brazil). J. Mar. Biol. Assoc. UK 2003, 83, 425–432. [Google Scholar] [CrossRef]
- Schmidt, E.W.; Donia, M.S. Life in cellulose houses: Symbiotic bacterial biosynthesis of ascidian drugs and drug leads. Curr. Opin. Biotech. 2010, 21, 827–833. [Google Scholar] [CrossRef] [PubMed]
- Palanisamy, S.K.; Rajendran, N.M.; Marino, A. Natural products diversity of marine ascidians (Tunicates; Ascidiacea) and successful drugs in clinical development. Nat. Prod. Bioprospect. 2017, 7, 1–111. [Google Scholar] [CrossRef] [PubMed]
- Mehbub, M.F.; Lei, J.; Franco, C.; Zhang, W. Marine sponge derived natural products between 2001 and 2010: Trends and opportunities for discovery of bioactives. Mar. Drugs 2014, 12, 4539–4577. [Google Scholar] [CrossRef]
- Molloy, E.M.; Hertweck, C. Antimicrobial discovery inspired by ecological interactions. Curr. Opin. Microbiol. 2017, 39, 121–127. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, E.W. The secret to a successful relationship: Lasting chemistry between ascidians and their symbiotic bacteria. Invertebr. Biol. 2015, 134, 88–102. [Google Scholar] [CrossRef]
- Steinert, G.; Taylor, M.W.; Schupp, P.J. Diversity of actinobacteria associated with the marine ascidian Eudistoma toealensis. Mar. Biotechnol. 2015, 17, 377–385. [Google Scholar] [CrossRef] [PubMed]
- Evans, J.S.; Erwin, P.M.; Shenkar, N.; Lopez-Legentil, S. Introduced ascidians harbor highly diverse and host-specific symbiotic microbial assemblages. Sci. Rep. 2017, 7, 11033–11043. [Google Scholar] [CrossRef]
- Pita, L.; Rix, L.; Slaby, B.M.; Franke, A.; Hentschel, U. The sponge holobiont in a changing ocean: From microbes to ecosystems. Microbiome 2018, 6, 2–18. [Google Scholar] [CrossRef] [PubMed]
- Bauermeister, A.; Branco, P.C.; Furtado, L.C.; Jimenez, P.C.; Costa-Lotufo, L.V.; da Cruz Lotufo, T.M. Tunicates: A model organism to investigate the effects of associated-microbiota on the production of pharmaceuticals. Drug Discov. Today Dis. Models 2019. [Google Scholar] [CrossRef]
- Chen, L.; Hu, J.S.; Xu, J.L.; Shao, C.L.; Wang, G.Y. Biological and chemical diversity of ascidian-associated microorganisms. Mar. Drugs 2018, 16, 362. [Google Scholar] [CrossRef]
- Andryukov, B.; Mikhailov, V.; Besednova, N. The Biotechnological Potential of Secondary Metabolites from Marine Bacteria. J. Mar. Sci. Eng. 2019, 7, 176. [Google Scholar] [CrossRef]
- Ferreira, E.G.; Torres, M.D.M.; da Silva, A.B.; Colares, L.L.F.; Pires, K.; Lotufo, T.M.C.; Silveira, E.R.; Pessoa, O.D.L.; Costa-Lotufo, L.V.; Jimenez, P.C. Prospecting anticancer compounds in actinomycetes recovered from the sediments of Saint Peter and Saint Paul’s Archipelago, Brazil. Chem. Biodivers. 2016, 13, 1149–1157. [Google Scholar] [CrossRef]
- Prieto-Davo, A.; Dias, T.; Gomes, S.E.; Rodrigues, S.; Parera-Valadezl, Y.; Borralho, P.M.; Pereira, F.; Rodrigues, C.M.P.; Santos-Sanches, I.; Gaudencio, S.P. The Madeira Archipelago as a significant source of marine-derived actinomycete diversity with anticancer and antimicrobial potential. Front. Microbiol. 2016, 7, 1594. [Google Scholar] [CrossRef] [PubMed]
- Maldonado, L.A.; Stach, J.E.M.; Pathom-aree, W.; Ward, A.C.; Bull, A.T.; Goodfellow, M. Diversity of cultivable actinobacteria in geographically widespread marine sediments. Anton. Van Leeuw. 2005, 87, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Maldonado, L.A.; Fragoso-Yanez, D.; Perez-Garcia, A.; Rosellon-Druker, J.; Quintana, E. Actinobacterial diversity from marine sediments collected in Mexico. Anton. Van Leeuw. 2009, 95, 111–120. [Google Scholar] [CrossRef]
- Becerril-Espinosa, A.; Freel, K.C.; Jensen, P.R.; Soria-Mercado, I.E. Marine Actinobacteria from the Gulf of California: Diversity, abundance and secondary metabolite biosynthetic potential. Anton. Van Leeuw. 2013, 103, 809–819. [Google Scholar] [CrossRef]
- Lackner, G.; Peters, E.E.; Helfrich, E.J.N.; Piel, J. Insights into the lifestyle of uncultured bacterial natural product factories associated with marine sponges. Proc. Natl. Acad. Sci. USA 2017, 114, E347–E356. [Google Scholar] [CrossRef]
- Bauermeister, A.; Velasco-Alzate, K.; Dias, T.; Macedo, H.; Ferreira, E.G.; Jimenez, P.C.; Lotufo, T.M.C.; Lopes, N.P.; Gaudencio, S.P.; Costa-Lotufo, L.V. Metabolomic fingerprinting of Salinispora from Atlantic oceanic islands. Front. Microbiol. 2018, 9, 3021. [Google Scholar] [CrossRef]
- Kemung, H.M.; Tan, L.T.H.; Khan, T.M.; Chan, K.G.; Pusparajah, P.; Goh, B.H.; Lee, L.H. Streptomyces as a prominent resource of future anti-MRSA drugs. Front. Microbiol. 2018, 9, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Ser, H.L.; Tan, L.T.H.; Law, J.W.F.; Chan, K.G.; Duangjai, A.; Saokaew, S.; Pusparajah, P.; Ab Mutalib, N.S.; Khan, T.M.; Goh, B.H.; et al. Focused review: Cytotoxic and antioxidant potentials of mangrove-derived Streptomyces. Front. Microbiol. 2017, 8, 1–11. [Google Scholar] [CrossRef]
- Brunetti, A.E.; Carnevale Neto, F.; Vera, M.C.; Taboada, C.; Pavarini, D.P.; Bauermeister, A.; Lopes, N.P. An integrative omics perspective for the analysis of chemical signals in ecological interactions. Chem. Soc. Rev. 2018, 47, 1574–1591. [Google Scholar] [CrossRef]
- Demarque, D.P.; Crotti, A.E.; Vessecchi, R.; Lopes, J.L.; Lopes, N.P. Fragmentation reactions using electrospray ionization mass spectrometry: An important tool for the structural elucidation and characterization of synthetic and natural products. Nat. Prod. Rep. 2016, 33, 432–455. [Google Scholar] [CrossRef]
- Da Silva, R.R.; Wang, M.; Nothias, L.F.; van der Hooft, J.J.J.; Caraballo-Rodríguez, A.M.; Fox, E.; Balunas, M.J.; Klassen, J.L.; Lopes, N.P.; Dorrestein, P.C. Propagating annotations of molecular networks using in silico fragmentation. PLoS Comput. Biol. 2018, 14, e1006089. [Google Scholar] [CrossRef]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.M.; Liang, X.A.; Kong, Y.; Jia, B. Structural diversity and biological activities of indole diketopiperazine alkaloids from fungi. J. Agric. Food Chem. 2016, 64, 6659–6671. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, M.M.; Andrade, M.D.; Bauermeister, A.; Merfa, M.V.; Forim, M.R.; Fernandes, J.B.; Vieira, P.C.; da Silva, M.F.D.F.; Lopes, N.P.; Machado, M.A.; et al. A simple defined medium for the production of true Diketopiperazines in Xylella fastidiosa and their identification by ultra-fast liquid chromatography-electrospray ionization ion trap mass spectrometry. Molecules 2017, 22, 985. [Google Scholar] [CrossRef] [PubMed]
- Rabindran, S.K.; He, H.Y.; Singh, M.; Brown, E.; Collins, K.I.; Annable, T.; Greenberger, L.M. Reversal of a novel multidrug resistance mechanism in human colon carcinoma cells by fumitremorgin C. Cancer Res. 1998, 58, 5850–5858. [Google Scholar] [PubMed]
- Bofinger, M.R.; de Sousa, L.S.; Fontes, J.E.N.; Marsaioli, A.J. Diketopiperazines as cross-communication quorum-sensing signals between Cronobacter sakazakii and Bacillus cereus. ACS Omega 2017, 2, 1003–1008. [Google Scholar] [CrossRef] [PubMed]
- Alfarisi, O.; Alghamdi, W.A.; Al-Shaer, M.H.; Dooley, K.E.; Peloquin, C. A Rifampin vs. rifapentine: What is the preferred rifamycin for tuberculosis? Exp. Rev. Clin. Pharm. 2017, 10, 1027–1036. [Google Scholar] [CrossRef] [PubMed]
- Meena, K.R.; Kanwar, S.S. Lipopeptides as the antifungal and antibacterial agents: Applications in food safety and therapeutics. Biomed. Res. Int. 2015, 2015, 1–9. [Google Scholar] [CrossRef]
- Vazquez-Laslop, N.; Mankin, A.S. How macrolide antibiotics work. Trends Biochem. Sci. 2018, 43, 668–684. [Google Scholar] [CrossRef]
- Katayama, N.; Fukusumi, S.; Funabashi, Y.; Iwahi, T.; Ono, H. TAN-1057 A-D, new antibiotics with potent antibacterial activity against methicillin-resistant Staphylococcus aureus. Taxonomy, fermentation and biological activity. J. Antibiot. 1993, 46, 606–613. [Google Scholar] [CrossRef]
- Frlan, R.; Gobec, S. Inhibitors of cathepsin B. Curr. Med. Chem. 2006, 13, 2309–2327. [Google Scholar] [PubMed]
- Li, T.; Christensen, S.D.; Frankel, P.H.; Margolin, K.A.; Agarwala, S.S.; Luu, T.; Mack, P.C.; Lara, P.N., Jr.; Gandara, D.R. A phase II study of cell cycle inhibitor UCN-01 in patients with metastatic melanoma: A California Cancer Consortium trial. Investig. New Drugs 2012, 30, 741–748. [Google Scholar] [CrossRef] [PubMed]
- Yadav, S.S.; Prasad, C.B.; Prasad, S.B.; Pandey, L.K.; Singh, S.; Pradhan, S.; Narayan, G. Anti-tumor activity of staurosporine in the tumor microenvironment of cervical cancer: An in vitro study. Life Sci. 2015, 133, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Jimenez, P.C.; Wilke, D.V.; Ferreira, E.G.; Takeara, R.; de Moraes, M.O.; da Silveira, E.R.; da Cruz Lotufo, T.M.; Lopes, N.P.; Costa-Lotufo, L.V. Structure elucidation and anticancer activity of 7-oxostaurosporine derivatives from the Brazilian endemic tunicate Eudistoma vannamei. Mar. Drugs 2012, 10, 1092–1102. [Google Scholar] [CrossRef] [PubMed]
- Andréo, M.A.; Jimenez, P.C.; Siebra, J.B.; Costa-Lotufo, L.V.; Vessecchi, R.; Niehues, M.; Lopes, J.L.; Lopes, N.P. Systematic UPLC-ESI-MS/MS study on the occurrence of staurosporine and derivatives in associated marine microorganisms from Eudistoma vannamei. J. Braz. Chem. Soc. 2012, 23, 335–343. [Google Scholar] [CrossRef]
- Williams, P.G.; Asolkar, R.N.; Kondratyuk, T.; Pezzuto, J.M.; Jensen, P.R.; Fenical, W. Saliniketals A and B, bicyclic polyketides from the marine actinomycete Salinispora arenicola. J. Nat. Prod. 2007, 70, 83–88. [Google Scholar] [CrossRef]
- Hufsky, F.; Scheubert, K.; Bocker, S. New kids on the block: Novel informatics methods for natural product discovery. Nat. Prod. Rep. 2014, 31, 807–817. [Google Scholar] [CrossRef]
- Hufsky, F.; Bocker, S. Mining molecular structure databases: Identification of small molecules based on fragmentation mass spectrometry data. Mass Spectrom. Rev. 2017, 36, 624–633. [Google Scholar] [CrossRef]
- Mohimani, H.; Gurevich, A.; Shlemov, A.; Mikheenko, A.; Korobeynikov, A.; Cao, L.; Shcherbin, E.; Nothias, L.F.; Dorrestein, P.C.; Pevzner, P.A. Dereplication of microbial metabolites through database search of mass spectra. Nat. Commun. 2018, 9, 4035. [Google Scholar] [CrossRef]
- Wan, Z.Y.; Fang, W.; Shi, L.Q.; Wang, K.M.; Zhang, Y.N.; Zhang, Z.G.; Wu, Z.Y.; Yang, Z.W.; Gu, Y.C. Novonestmycins A and B, two new 32-membered bioactive macrolides from Streptomyces phytohabitans HBERC-20821. J. Antibiot. 2015, 68, 185–190. [Google Scholar] [CrossRef]
- Crowe, M.C.; Brodbelt, J.S.; Goolsby, B.J.; Hergenrother, P. Characterization of erythromycin analogs by collisional activated dissociation and infrared multiphoton dissociation in a quadrupole ion trap. J. Am. Soc. Mass Spectr. 2002, 13, 630–649. [Google Scholar] [CrossRef]
- Wills, R.H.; Tosin, M.; O’Connor, P.B. Structural characterization of polyketides using high mass accuracy tandem mass spectrometry. Anal. Chem. 2012, 84, 8863–8870. [Google Scholar] [CrossRef] [PubMed]
- Gontang, E.A.; Fenical, W.; Jensen, P.R. Phylogenetic diversity of gram-positive bacteria cultured from marine sediments. Appl. Environ. Microbiol. 2007, 73, 3272–3282. [Google Scholar] [CrossRef] [PubMed]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Met. 1983, 65, 55–63. [Google Scholar] [CrossRef]
- Kumar, V.; Bisht, G.S.; Institu, S.B.S.P.G. An improved method for isolation of genomic DNA from filamentous actinomycetes. J. Sci. Eng. Technol. Mgt. 2010, 2, 10–13. [Google Scholar]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016, 44, W242–W245. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]




© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Velasco-Alzate, K.Y.; Bauermeister, A.; Tangerina, M.M.P.; Lotufo, T.M.C.; Ferreira, M.J.P.; Jimenez, P.C.; Padilla, G.; Lopes, N.P.; Costa-Lotufo, L.V. Marine Bacteria from Rocas Atoll as a Rich Source of Pharmacologically Active Compounds. Mar. Drugs 2019, 17, 671. https://doi.org/10.3390/md17120671
Velasco-Alzate KY, Bauermeister A, Tangerina MMP, Lotufo TMC, Ferreira MJP, Jimenez PC, Padilla G, Lopes NP, Costa-Lotufo LV. Marine Bacteria from Rocas Atoll as a Rich Source of Pharmacologically Active Compounds. Marine Drugs. 2019; 17(12):671. https://doi.org/10.3390/md17120671
Chicago/Turabian StyleVelasco-Alzate, Karen Y., Anelize Bauermeister, Marcelo M. P. Tangerina, Tito M. C. Lotufo, Marcelo J. P. Ferreira, Paula C. Jimenez, Gabriel Padilla, Norberto P. Lopes, and Letícia V. Costa-Lotufo. 2019. "Marine Bacteria from Rocas Atoll as a Rich Source of Pharmacologically Active Compounds" Marine Drugs 17, no. 12: 671. https://doi.org/10.3390/md17120671
APA StyleVelasco-Alzate, K. Y., Bauermeister, A., Tangerina, M. M. P., Lotufo, T. M. C., Ferreira, M. J. P., Jimenez, P. C., Padilla, G., Lopes, N. P., & Costa-Lotufo, L. V. (2019). Marine Bacteria from Rocas Atoll as a Rich Source of Pharmacologically Active Compounds. Marine Drugs, 17(12), 671. https://doi.org/10.3390/md17120671







