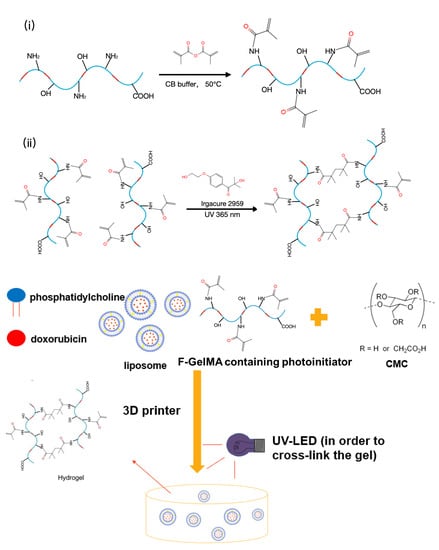
Fabrication of 3D-Printed Fish-Gelatin-Based Polymer Hydrogel Patches for Local Delivery of PEGylated Liposomal Doxorubicin
Abstract
1. Introduction
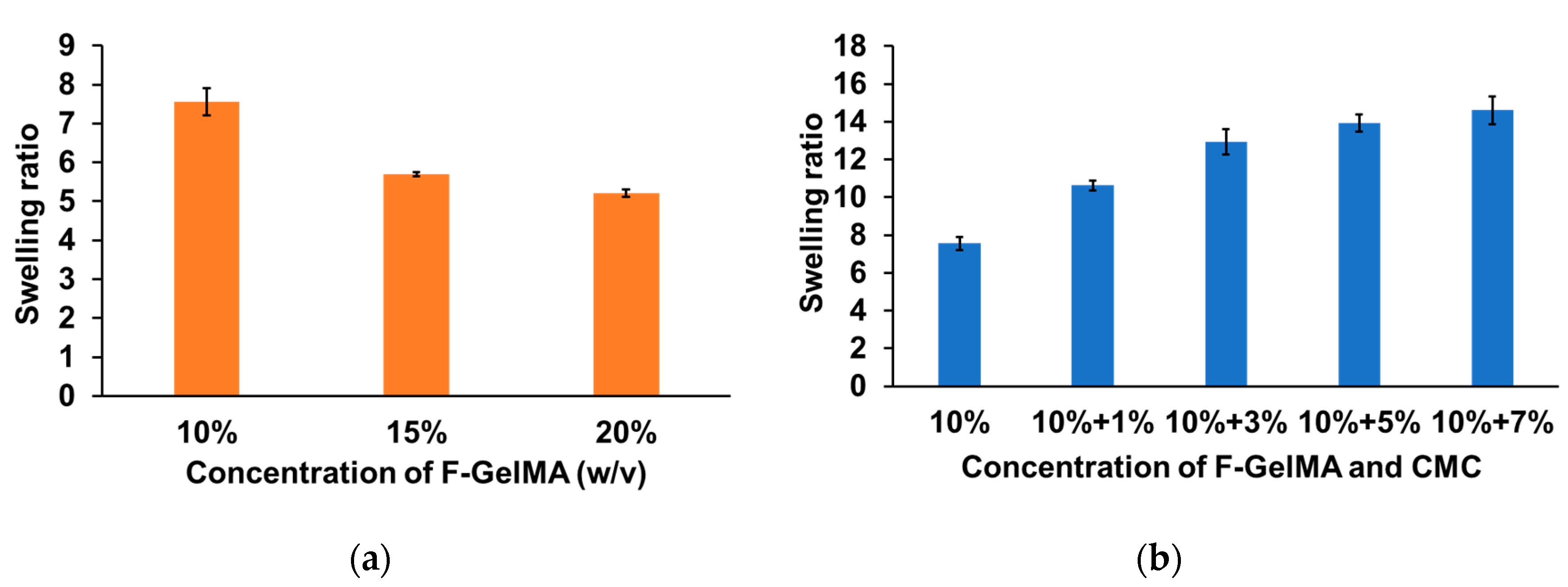
2. Results and Discussion
3. Materials and Methods
3.1. Materials
3.2. Synthesis of F-GelMA
3.3. Evaluation of the Degree of Methacrylation of F-GelMA
3.4. Hydrogel Swelling Test after Photopolymerization
3.5. Viscosity Test of Drug Formulation for Use as a Printer Ink
3.6. Preparation of DOX-Loaded PEGylated Liposomes
3.7. Encapsulation Efficiency of DOX into the Liposomes
3.8. Stability Test of Liposomes Following Photopolymerization of the Hydrogel
3.9. 3D Bioprinting
3.10. In Vitro Release of DOX-Loaded Liposomes from 3D-Printed Patches
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Bozzuto, G.; Molinari, A. Liposomes as nanomedical devices. Int. J. Nanomed. 2015, 10, 975–999. [Google Scholar] [CrossRef] [PubMed]
- Allen, T.M.; Cullis, P.R. Liposomal drug delivery systems: From concept to clinical applications. Adv. Drug Deliv. Rev. 2013, 65, 36–48. [Google Scholar] [CrossRef] [PubMed]
- Tran, S.; DeGiovanni, P.J.; Piel, B.; Rai, P. Cancer nanomedicine: A review of recent success in drug delivery. Clin. Transl. Med. 2017, 6, 44. [Google Scholar] [CrossRef]
- Barenholz, Y. Doxil®—The first FDA-approved nano-drug: Lessons learned. J. Control. Release 2013, 160, 117–134. [Google Scholar] [CrossRef] [PubMed]
- Gabizon, A.; Shmeeda, H.; Barenholz, Y. Pharmacokinetics of pegylated liposomal doxorubicin: Review of animal and human studies. Clin. Pharmacokinet. 2003, 42, 419–436. [Google Scholar] [CrossRef]
- Abu Lila, A.S.; Ishida, T. Liposomal delivery systems: Design optimization and current applications. Biol. Pharm. Bull. 2017, 40, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Chakroun, R.W.; Zhang, P.; Lin, R.; Schiapparelli, P.; Quinones-Hinojosa, A.; Cui, H. Nanotherapeutic systems for local treatment of brain tumors. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2018, 10, e1479. [Google Scholar] [CrossRef] [PubMed]
- Mazaheri, M.; Eslahi, N.; Ordikhani, F.; Tamjid, E.; Simchi, A. Nanomedicine applications in orthopedic medicine: State of the art. Int. J. Nanomed. 2015, 10, 6039–6054. [Google Scholar]
- Talebian, S.; Foroughi, J.; Wade, S.J.; Vine, K.L.; Dolatshahi-Pirouz, A.; Mehrali, M.; Conde, J.; Wallace, G.G. Biopolymers for antitumor implantable drug delivery systems: Recent advances and future outlook. Adv. Mater. 2018, 30, 1706665. [Google Scholar] [CrossRef]
- Ventola, C.L. Medical applications for 3D printing: Current and projected uses. Pharm. Ther. 2014, 39, 704–711. [Google Scholar]
- Tack, P.; Victor, J.; Gemmel, P.; Annemans, L. 3D-printing techniques in a medical setting. Biomed. Eng. Online 2016, 15, 115. [Google Scholar] [CrossRef] [PubMed]
- Jamroz, W.; Szafraniec, J.; Kurek, M.; Jachowicz, R. 3D printing in pharmaceutical and medical applications—Recent achievements and challenges. Pharm. Res. 2018, 35, 176. [Google Scholar] [CrossRef] [PubMed]
- Aprecia Pharmaceuticals. Available online: https://www.aprecia.com/ (accessed on 24 March 2020).
- Alhnan, M.A.; Okwuosa, T.C.; Sadia, M.; Wan, K.W.; Ahmed, W.; Arafat, B. Emergence of 3D printed dosage forms: Opportunities and challenges. Pharm. Res. 2016, 33, 1817–1832. [Google Scholar] [CrossRef] [PubMed]
- Witowski, J.; Sitkowski, M.; Zuzak, T.; Coles-Black, J.; Chuen, J.; Major, P.; Pdziwiatr, M. From ideas to long-term studies: 3D printing clinical trials review. Int. J. Comput. Assist. Radiol. Surg. 2018, 13, 1473–1478. [Google Scholar] [CrossRef]
- Derakhshanfar, S.; Mbeleck, R.; Xu, K.; Zhang, X.; Zhong, W.; Xing, M. 3D bioprinting for biomedical devices and tissue engineering: A review of recent trends and advances. Bioact. Mater. 2018, 3, 144–156. [Google Scholar] [CrossRef]
- Chimene, D.; Lennox, K.K.; Kaunas, R.R.; Gaharwar, A.K. Advanced bioinks for 3D printing: A materials science perspective. Ann. Biomed. Eng. 2016, 44, 2090–2102. [Google Scholar] [CrossRef]
- Ouyang, L.; Highley, C.B.; Sun, W.; Burdick, J.A. A generalizable strategy for the 3D bioprinting of hydrogels from nonviscous photo-crosslinkable Inks. Adv. Mater. 2017, 29, 1604983. [Google Scholar] [CrossRef]
- Yi, H.G.; Choi, Y.J.; Kang, K.S.; Hong, J.M.; Pati, R.G.; Park, M.N.; Shim, I.K.; Lee, C.M.; Kim, S.C.; Cho, D.W. A 3D-printed local drug delivery patch for pancreatic cancer growth suppression. J. Control. Release 2016, 238, 231–241. [Google Scholar] [CrossRef]
- Yue, K.; Trujillo-de Santiago, G.; Alvarez, M.M.; Tamayol, A.; Annabi, N.; Khademhosseini, A. Synthesis, properties, and biomedical applications of gelatin methacryloyl (GelMA) hydrogels. Biomaterials 2015, 73, 254–271. [Google Scholar] [CrossRef]
- Hassanzadeh, P.; Kazemzadeh-Narbat, M.; Rosenzweig, R.; Zhang, X.; Khademhosseini, A.; Annabi, N.; Rolandi, M. Ultrastrong and Flexible Hybrid Hydrogels Based on Solution Self-Assembly of Chitin Nanofibers in Gelatin Methacryloyl (GelMA). J. Mater. Chem. B 2016, 4, 2539–2543. [Google Scholar] [CrossRef]
- Karim, A.A.; Bhat, R. Fish gelatin: Properties, challenges, and prospects as an alternative to mammalian gelatins. Food Hydrocoll. 2009, 23, 563–576. [Google Scholar] [CrossRef]
- Yoon, H.J.; Shin, S.R.; Cha, J.M.; Lee, S.H.; Kim, J.H.; Do, J.T.; Song, H.; Bae, H. Cold water fish gelatin methacryloyl hydrogel for tissue engineering application. PLoS ONE 2016, 11, e0163902. [Google Scholar] [CrossRef] [PubMed]
- Zaupa, A.; Byres, N.; Dal Zovo, C.; Acevedo, C.A.; Angelopoulos, I.; Terraza, C.; Nestle, N.; Abarzúa-Illanes, P.N.; Quero, F.; Díaz-Calderón, P.; et al. Cold-adaptation of a methacrylamide gelatin towards the expansion of the biomaterial toolbox for specialized functionalities in tissue engineering. Mater. Sci. Eng. C Mater. Biol. Appl. 2019, 102, 373–390. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Kim, G.J.; Kang, M.G.; Lee, J.K.; Seo, J.W.; Do, J.T.; Hong, K.; Cha, J.M.; Shin, S.R.; Bae, H. Marine biomaterial-based bioinks for generating 3D printed tissue constructs. Mar. Drugs 2018, 16, 484. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Tian, Z.; Menard, F.; Kim, K. Comparative study of gelatin methacrylate hydrogels from different sources for biofabrication applications. Biofabrication 2017, 9, 0444101. [Google Scholar] [CrossRef] [PubMed]
- Kang, M.G.; Lee, M.Y.; Cha, J.M.; Lee, J.K.; Lee, S.C.; Kim, J.; Hwang, Y.S.; Bae, H. Nanogels derived from fish gelatin: Application to drug delivery system. Mar. Drugs 2019, 17, 246. [Google Scholar] [CrossRef] [PubMed]
- Shirahama, H.; Lee, B.H.; Tan, L.P.; Cho, N.J. Precise tuning of facile one-pot gelatin methacryloyl (GelMA) synthesis. Sci. Rep. 2016, 6, 31036. [Google Scholar] [CrossRef]
- Habeeb, A.F.S.A. Determination of free amino groups in proteins by trinitrobenzenesulfonic acid. Anal. Biochem. 1966, 14, 328–336. [Google Scholar] [CrossRef]
- Haug, I.J.; Draget, K.I.; Smidsrød, O. Physical and rheological properties of fish gelatin compared to mammalian gelatin. Food Hydrocoll. 2004, 18, 203–213. [Google Scholar] [CrossRef]
- Kamel, S.; Ali, N.; Jahangir, K.; Shah, S.M.; El-Gendy, A.A. Pharmaceutical significance of cellulose: A review. Express Polym. Lett. 2008, 2, 78–87. [Google Scholar] [CrossRef]
- Tagami, T.; Yoshimura, N.; Goto, E.; Noda, T.; Ozeki, T. Fabrication of muco-adhesive oral films by the 3D printing of hydroxypropyl methylcellulose-based catechin-loaded formulations. Biol. Pharm. Bull. 2019, 42, 1898–1905. [Google Scholar] [CrossRef] [PubMed]
- Nichol, J.W.; Koshy, S.T.; Bae, H.; Hwang, C.M.; Yamanlar, S.; Khademhosseini, A. Cell-laden microengineered gelatin methacrylate hydrogels. Biomaterials 2010, 31, 5536–5544. [Google Scholar] [CrossRef] [PubMed]
- Ganji, F.; Vasheghani, H.S.; Vasheghani, F.E. Theoretical description of hydrogel swelling: A review. Iran. Polym. J. 2010, 19, 375–398. [Google Scholar]
- Cheng, R.; Yan, Y.; Liu, H.; Chen, H.; Pan, G.; Deng, L.; Cui, W. Mechanically enhanced lipo-hydrogel with controlled release of multi-type drugs for bone regeneration. Appl. Mater. Today 2018, 12, 294–308. [Google Scholar] [CrossRef]
- Chen, Y.X.; Cain, B.; Soman, P. Gelatin methacrylate-alginate hydrogel with tunable viscoelastic properties. AIMS Mater. Sci. 2017, 4, 363–369. [Google Scholar] [CrossRef]
- Martinez, P.R.; Goyanes, A.; Basit, A.W.; Gaisford, S. Influence of geometry on the drug release profiles of stereolithographic (SLA) 3D-printed tablets. AAPSPharmSciTech 2018, 19, 3355–3361. [Google Scholar] [CrossRef]
- Tagami, T.; Nagata, N.; Hayashi, N.; Ogawa, E.; Fukushige, K.; Sakai, N.; Ozeki, T. Defined drug release from 3D-printed composite tablets consisting of drug-loaded polyvinylalcohol and a water-soluble or water-insoluble polymer filler. Int. J. Pharm. 2018, 543, 361–367. [Google Scholar] [CrossRef]
- Lee, B.H.; Shirahama, H.; Cho, N.J.; Tan, L.P. Efficient and controllable synthesis of highly substituted gelatin methacrylamide for mechanically stiff hydrogels. RSC Adv. 2015, 5, 106094–106097. [Google Scholar] [CrossRef]
- Benton, J.A.; DeForest, C.A.; Vivekanandan, V.; Anseth, K.S. Photocrosslinking of gelatin macromers to synthesize porous hydrogels that promote valvular interstitial cell function. Tissue Eng. Part A 2009, 15, 3221–3230. [Google Scholar] [CrossRef]
- Tagami, T.; Kubota, M.; Ozeki, T. Effective remote loading of doxorubicin into DPPC/Poloxamer 188 hybrid liposome to retain thermosensitive property and the assessment of carrier-based acute cytotoxicity for pulmonary administration. J. Pharm. Sci. 2015, 104, 3824–3832. [Google Scholar] [CrossRef]
- Song, M.; Wang, J.; Lei, J.; Peng, G.; Zhang, W.; Zhang, Y.; Yin, M.; Li, J.; Liu, Y.; Wei, X.; et al. Preparation and evaluation of liposomes co-loaded with doxorubicin, phospholipase D inhibitor 5-fluoro-2-indolyl deschlorohalopemide (FIPI) and D-alpha tocopheryl acid succinate (α-TOS) for anti-metastasis. Nanoscale Res. Lett. 2019, 14, 138. [Google Scholar] [CrossRef] [PubMed]
- Tagami, T.; Ernsting, M.J.; Li, S.D. Efficient tumor regression by a single and low dose treatment with a novel and enhanced formulation of thermosensitive liposomal doxorubicin. J. Control. Release 2011, 152, 303–309. [Google Scholar] [CrossRef] [PubMed]






| 3D Geometries | Nozzle Diameter | Pressure |
|---|---|---|
| Cylinder | 22 G | 65 |
| Torus | 22 G | 65 |
| Gridlines | 27 G | 70 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, J.; Tagami, T.; Ozeki, T. Fabrication of 3D-Printed Fish-Gelatin-Based Polymer Hydrogel Patches for Local Delivery of PEGylated Liposomal Doxorubicin. Mar. Drugs 2020, 18, 325. https://doi.org/10.3390/md18060325
Liu J, Tagami T, Ozeki T. Fabrication of 3D-Printed Fish-Gelatin-Based Polymer Hydrogel Patches for Local Delivery of PEGylated Liposomal Doxorubicin. Marine Drugs. 2020; 18(6):325. https://doi.org/10.3390/md18060325
Chicago/Turabian StyleLiu, Jin, Tatsuaki Tagami, and Tetsuya Ozeki. 2020. "Fabrication of 3D-Printed Fish-Gelatin-Based Polymer Hydrogel Patches for Local Delivery of PEGylated Liposomal Doxorubicin" Marine Drugs 18, no. 6: 325. https://doi.org/10.3390/md18060325
APA StyleLiu, J., Tagami, T., & Ozeki, T. (2020). Fabrication of 3D-Printed Fish-Gelatin-Based Polymer Hydrogel Patches for Local Delivery of PEGylated Liposomal Doxorubicin. Marine Drugs, 18(6), 325. https://doi.org/10.3390/md18060325