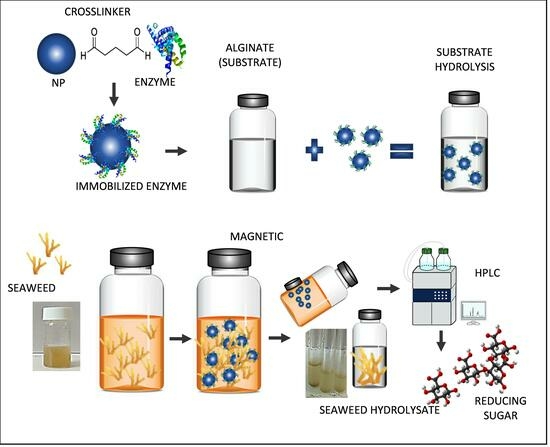
Production of Alginate Oligosaccharides (AOSs) Using Enhanced Physicochemical Properties of Immobilized Alginate Lyase for Industrial Application
Abstract
:1. Introduction
2. Results and Discussion
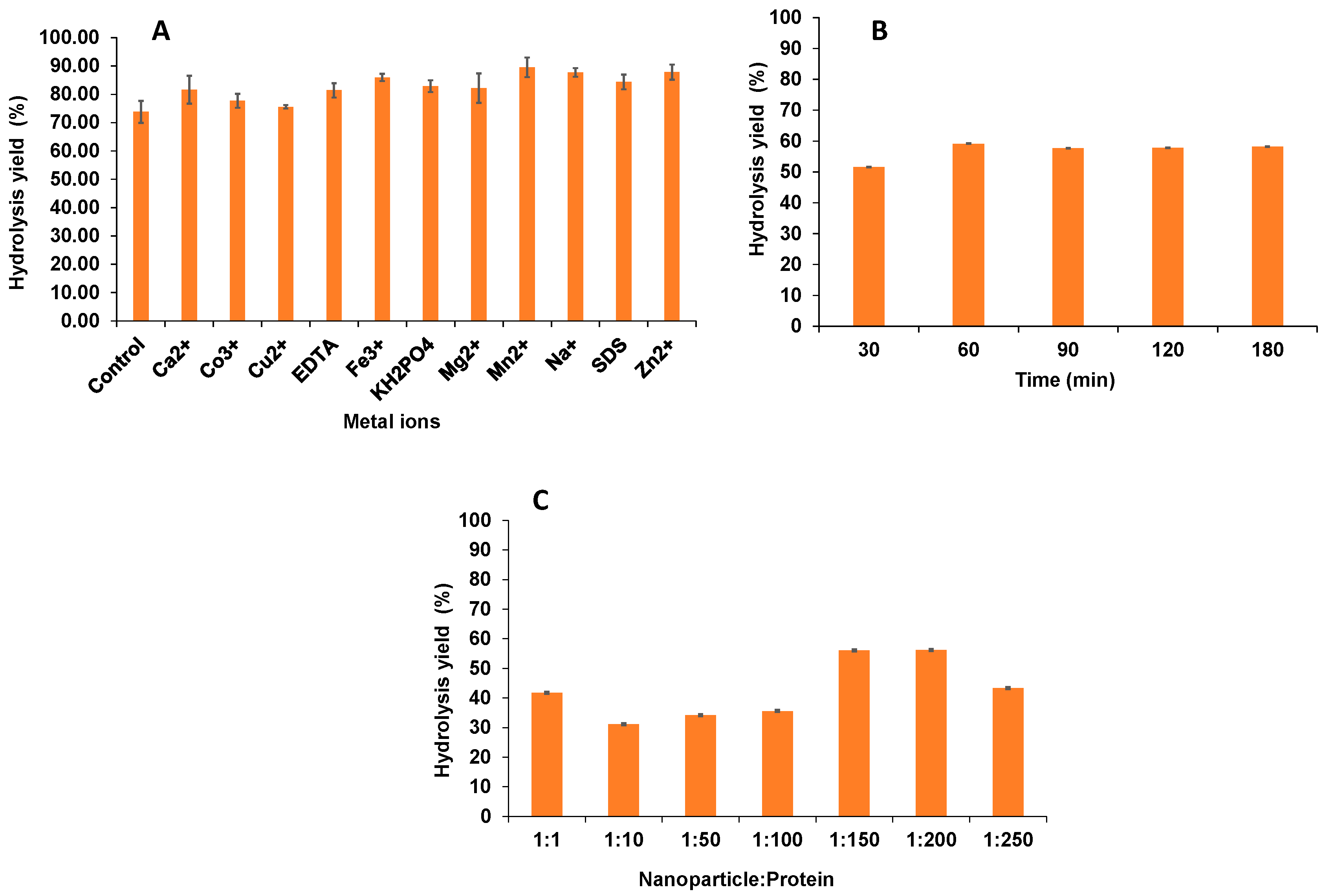
2.1. Effect of Metal Ions on Enzymatic Activity and Binding Efficiency of Immobilization
2.2. Characterization Studies
2.3. Enzyme Activity at Varying pH and Temperature
2.4. Kinetic Study on Immobilized and Free Enzyme
2.5. Thermal Stability and Storage Stability of Alginate Lyase
2.6. Reusability of Immobilized Enzyme
2.7. Enzymatic Saccharification of Raw Seaweed Biomass
2.8. High-Performance Liquid Chromatography (HPLC) Analysis
2.9. Antioxidant Assay
3. Materials and Methods
3.1. Enzyme Assay
3.2. Effect of Different Metal Ions on Enzymatic Activity
3.3. Immobilization of Alginate Lyase
3.4. Characterization of Immobilized Alginate Lyase
3.5. Temperature and pH Optimization
3.6. Determination of Enzyme Kinetics
3.7. Thermostability and Storage Stability
3.8. Reusability
3.9. Enzyme Saccharification of Seaweed Biomass
3.10. Antioxidant Assay
3.11. High-Performance Liquid Chromatography Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, Q.; Hu, F.; Wang, M.; Zhu, B.; Ni, F.; Yao, Z. Elucidation of degradation pattern and immobilization of a novel alginate lyase for preparation of alginate oligosaccharides. Int. J. Biol. Macromol. 2020, 146, 579–587. [Google Scholar] [CrossRef]
- Wong, T.Y.; Preston, L.A.; Schiller, N.L. Alginate lyase: Review of major sources and enzyme characteristics, structure-function analysis, biological roles, and applications. Annu. Rev. Microbiol. 2000, 54, 289–340. [Google Scholar] [CrossRef] [PubMed]
- Batista, P.S.P.; de Morais, A.M.M.B.; Pintado, M.M.E. Alginate: Pharmaceutical and Medical Applications. In Extracellular Sugar-Based Biopolymers Matrices; Ephraim, C., Hans, M., Eds.; Springer: Cham, Germany, 2019; pp. 649–691. [Google Scholar]
- Xing, M.; Cao, Q.; Wang, Y.; Xiao, H.; Zhao, J.; Zhang, Q.; Ji, A.; Song, S. Advances in research on the bioactivity of alginate oligosaccharides. Mar. Drugs 2020, 18, 144. [Google Scholar] [CrossRef]
- Cheng, D.; Jiang, C.; Xu, J.; Liu, Z.; Mao, X. Characteristics and applications of alginate lyases: A review. Int. J. Biol. Macromol. 2020, 164, 1304–1320. [Google Scholar] [CrossRef] [PubMed]
- Peng, C.; Wang, Q.; Lu, D.; Han, W.; Li, F. A novel bifunctional endolytic alginate lyase with variable alginate-degrading modes and versatile monosaccharide-producing properties. Front. Microbiol. 2018, 9, 167. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Li, M.; Rauf, A.; Khalil, A.A.; Shan, Z.; Chen, C.; Rengasamy, K.R.R. Process and applications of alginate oligosaccharides with emphasis on health beneficial perspectives. Crit. Rev. Food Sci. Nutr. 2021, 63, 1–27. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Zhang, X.; Wu, L.; Li, H.; Chen, Y.; Lijun, L.; Ni, H.; Li, Q.; Hu, Y. Exolytic products of alginate by the immobilized alginate lyase confer antioxidant and antiapoptotic bioactivities in human umbilical vein endothelial cells. Carbohydr. Polym. 2021, 251, 16976. [Google Scholar] [CrossRef]
- Bie, J.; Sepodes, B.; Fernandes, P.C.B.; Ribeiro, M.H.L. Enzyme Immobilization and Co-Immobilization: Main Framework, Advances and Some Applications. Processes 2022, 10, 494. [Google Scholar] [CrossRef]
- Badoei-dalfard, A.; Tahami, A.; Karami, Z. Lipase immobilization on glutaraldehyde activated graphene oxide/chitosan/cellulose acetate electrospun nanofibrous membranes and its application on the synthesis of benzyl acetate. Colloids Surf. B Biointerfaces 2022, 209, 112151. [Google Scholar] [CrossRef]
- Patel, K.K.; Tripathi, M.; Pandey, N.; Agrawal, A.K.; Gade, S.; Anjum, M.M.; Tilak, R.; Singh, S. Alginate lyase immobilized chitosan nanoparticles of ciprofloxacin for the improved antimicrobial activity against the biofilm associated mucoid P. aeruginosa infection in cystic fibrosis. Int. J. Pharm. 2019, 563, 30–42. [Google Scholar] [CrossRef]
- Li, S.; Wang, Y.; Li, X.; Lee, B.S.; Jung, S.; Lee, M.S. Enhancing the thermo-stability and anti-biofilm activity of alginate lyase by immobilization on low molecular weight chitosan nanoparticles. Int. J. Mol. Sci. 2019, 20, 4565. [Google Scholar] [CrossRef] [PubMed]
- Mohapatra, B.R. Characterization of β-mannanase extracted from a novel Streptomyces species Alg-S25 immobilized on chitosan nanoparticles. Biotechnol. Biotechnol. Equip. 2021, 35, 150–161. [Google Scholar] [CrossRef]
- Magro, L.D.; Kornecki, J.F.; Klein, M.P.; Rodrigues, R.C.; Fernandez-Lafuente, R. Pectin lyase immobilization using the glutaraldehyde chemistry increases the enzyme operation range. Enzym. Microb. Technol. 2020, 132, 109397. [Google Scholar] [CrossRef] [PubMed]
- Joseph, J.E.; Mary, P.R.; Haritha, K.V.; Panwar, D.; Kapoor, M. Soluble and Cross-Linked Aggregated Forms of α-Galactosidase from Vigna mungo Immobilized on Magnetic Nanocomposites: Improved Stability and Reusability. Appl. Biochem. Biotechnol. 2021, 193, 238–256. [Google Scholar] [CrossRef] [PubMed]
- Verma, L.M.; Chaudhary, R.; Suzuki, T.; Barrow, C.; Puri, M. Immobilization of b-glucosidase on magnetic nanoparticle improves thermostability. Bioresour. Technol. 2013, 135, 2–6. [Google Scholar] [CrossRef]
- Puri, M.; Verma, M. Enzyme immobilization on nanomaterials for biofuel production. Trends Biotechnol. 2013, 31, 215–216. [Google Scholar] [CrossRef]
- Lu, D.; Zhang, Q.; Wang, S.; Guan, J.; Jiao, R.; Han, N.; Han, W.; Li, F. Biochemical characteristics and synergistic effect of two novel alginate lyases from Photobacterium sp. FC615. Biotechnol. Biofuels Bioprod. 2019, 12, 1–17. [Google Scholar] [CrossRef]
- Zhou, H.X.; Xu, S.S.; Yin, X.J.; Wang, F.L.; Li, Y. Characterization of a new bifunctional and cold-adapted polysaccharide lyase (PL) family 7 alginate lyase from Flavobacterium sp. Mar. Drugs 2020, 18, 388. [Google Scholar] [CrossRef]
- Ma, Y.; Li, J.; Zhang, X.-Y.; Ni, H.-D.; Wang, F.-B.; Wang, H.-Y.; Wang, Z.-P. Characterization of a New Intracellular Alginate Lyase with Metal Ions-Tolerant and pH-Stable Properties. Mar. Drugs 2020, 18, 416. [Google Scholar] [CrossRef]
- Boucelkha, A.; Petit, E.; Elboutachfaiti, R.; Molinié, R.; Amari, S.; Zaidi-Yahaoui, R. Production of guluronate oligosaccharide of alginate from brown algae Stypocaulon scoparium using an alginate lyase. J. Appl. Phycol. 2017, 29, 509–519. [Google Scholar] [CrossRef]
- Huang, L.; Zhou, J.; Li, X.; Peng, Q.; Lu, H.; Du, Y. Characterization of a new alginate lyase from newly isolated Flavobacterium sp. S20. J. Ind. Microbiol. Biotechnol. 2013, 40, 113–122. [Google Scholar] [CrossRef]
- Li, N.; Fu, X.; Xiao, M.; Wei, X.; Yang, M.; Liu, Z.; Mou, H. Enzymatic preparation of a low-molecular-weight polysaccharide rich in uronic acid from the seaweed Laminaria japonica and evaluation of its hypolipidemic effect in mice. Food Funct. 2020, 11, 2395–2405. [Google Scholar] [CrossRef]
- Sun, M.; Sun, C.; Li, T.; Li, K.; Yan, S.; Yin, H. Characterization of a novel bifunctional mannuronan C-5 epimerase and alginate lyase from Pseudomonas mendocina. sp. DICP-70. Int. J. Biol. Macromol. 2020, 150, 662–670. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Jiang, X.; Guan, H.; Wang, P. Preparation, purification and characterization of alginate oligosaccharides degraded by alginate lyase from Pseudomonas sp. HZJ 216. Carbohydr. Res. 2011, 346, 794–800. [Google Scholar] [CrossRef] [PubMed]
- Blanco-Cabra, N.; Paetzold, B.; Ferrar, T.; Mazzolini, R.; Torrents, E.; Serrano, L.; Lluch-Senar, M. Characterization of different alginate lyases for dissolving Pseudomonas aeruginosa biofilms. Sci. Rep. 2020, 10, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Hu, F.; Cao, S.; Li, Q.; Zhu, B.; Yao, Z. Construction and biochemical characterization of a novel hybrid alginate lyase with high activity by module recombination to prepare alginate oligosaccharides. Int. J. Biol. Macromol. 2021, 166, 1272–1279. [Google Scholar] [CrossRef]
- Zhu, B.-W.; Huang, L.-S.-X.; Tan, H.-D.; Qin, Y.-Q.; Du, Y.-G.; Yin, H. Characterization of a new endo-type polyM-specific alginate lyase from Pseudomonas sp. Biotechnol. Lett. 2015, 37, 409–415. [Google Scholar] [CrossRef] [PubMed]
- Mohapatra, B.R. Biocatalytic characteristics of chitosan nanoparticle-immobilized alginate lyase extracted from a novel Arthrobacter species AD-10. Biocatal. Agric. Biotechnol. 2020, 23, 101458. [Google Scholar] [CrossRef]
- Zhu, B.; Hu, F.; Yuan, H.; Sun, Y.; Yao, Z. Biochemical characterization and degradation pattern of a unique pH-stable PolyM-specific alginate lyase from newly isolated Serratia marcescens NJ-07. Mar. Drugs 2018, 16, 129. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.-H.; Chen, X.-L.; Wang, X.-F.; Zhang, X.-R.; Sun, X.-M.; Sun, M.-L.; Zhang, X.-Y.; Zhang, Y.-Z.; Zhang, Y.-Q.; Xu, F. Cost-effective production of alginate oligosaccharides from Laminaria japonica roots by Pseudoalteromonas agarivorans A3. Microb. Cell Factories 2023, 22, 179. [Google Scholar] [CrossRef]
- Sun, C.; Zhou, J.; Duan, G.; Yu, X. Hydrolyzing Laminaria japonica with a combination of microbial alginate lyase and cellulase. Bioresour. Technol. 2020, 311, 123548. [Google Scholar] [CrossRef] [PubMed]
- Xue, Z.; Sun, X.-M.; Chen, C.; Zhang, X.-Y.; Chen, X.-L.; Zhang, Y.-Z.; Fan, S.-J.; Xu, F. A novel alginate lyase: Identification, characterization, and potential application in alginate trisaccharide preparation. Mar. Drugs 2022, 20, 159. [Google Scholar] [CrossRef] [PubMed]
- Bisaria, V.S.; Ghose, T.K. Biodegradation of cellulosic materials: Substrates, microorganisms, enzymes and products. Enzym. Microb. Technol. 1981, 3, 90–104. [Google Scholar] [CrossRef]
- Abraham, R.E.; Verma, M.L.; Barrow, C.J.; Puri, M. Suitability of magnetic nanoparticle immobilised cellulases in enhancing enzymatic saccharification of pretreated hemp biomass. Biotechnol. Biofuels Bioprod. 2014, 7, 1–12. [Google Scholar] [CrossRef]
- Wang, Z.P.; Cao, M.; Li, B.; Ji, X.F.; Zhang, X.Y.; Zhang, Y.Q.; Wang, H.Y. Cloning, secretory expression and characterization of a unique pH-stable and cold-adapted alginate lyase. Mar. Drugs 2020, 18, 189. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, H.; Pal, R.; Guha, A.K.; Chatterjee, B.P. Isolation, Purification and Specificity of the Lectin from Pseudomonas Aeruginosa Bacteria (Habs Stain H8). In Proceedings of the IUB Symposium No. 144, The Seventh International Lectin Meeting, Bruxelles, Belgium, 18–23 August 1985; De Gruyter: Berlin, Germany, 1986; pp. 305–314. [Google Scholar]
- Chua, M.; Chan, K.; Hocking, T.J.; Williams, P.A.; Perry, C.J.; Baldwin, T.C. Methodologies for the extraction and analysis of konjac glucomannan from corms of Amorphophallus konjac K. Koch. Carbohydr. Polym. 2012, 87, 2202–2210. [Google Scholar] [CrossRef]
- Nawawi, N.N.; Hashim, Z.; Rahman, R.A.; Murad, A.M.A.; Bakar, F.D.A.; Illias, R.M. Entrapment of porous cross-linked enzyme aggregates of maltogenic amylase from Bacillus lehensis G1 into calcium alginate for maltooligosaccharides synthesis. Int. J. Biol. Macromol. 2020, 150, 80–89. [Google Scholar] [CrossRef]
- Abraham, R.E.; Su, P.; Puri, M.; Raston, C.L.; Zhang, W. Optimisation of biorefinery production of alginate, fucoidan and laminarin from brown seaweed Durvillaea potatorum. Algebra Res. 2019, 38, 101389. [Google Scholar] [CrossRef]
- Romanini, E.B.; Rodrigues, L.M.; Finger, A.; Chierrito, T.P.C.; Da Silva Scapim, M.R.; Madrona, G.S. Ultrasound assisted extraction of bioactive compounds from BRS Violet grape pomace followed by alginate-Ca2+ encapsulation. Food Chem. 2021, 338, 128101. [Google Scholar] [CrossRef]
- Zhuo, R.; Li, B.; Tian, S. Alginate oligosaccharide improves resistance to postharvest decay and quality of kiwifruit (Actinidia deliciosa ‘Bruno’). Hortic. Plant J. 2022, 8, 44–52. [Google Scholar] [CrossRef]
- Abraham, R.E.; Su, P.; Puri, M.; Raston, C.L.; Zhang, W. Release of encapsulated bioactives influenced by alginate viscosity under in-vitro gastrointestinal model. Inter. J Biol. Macromol. 2021, 170, 540–548. [Google Scholar] [CrossRef] [PubMed]
- Abraham, R.E. Bioprocessing of Hemp Hurd (Cannabis sativa) for Biofuel Production. Ph.D. Thesis, Deakin University, Melbourne, Australia, 2014. [Google Scholar]



| Parameters | Soluble Enzyme | Immobilized Enzyme |
|---|---|---|
| Km (mM) | 10.10 | 20 |
| Vmax (mg/mL/min) | 0.2 | 0.2 |
| Oligosaccharides | Soluble Enzyme (%, w/w) | Immobilized Enzyme (%, w/w) |
|---|---|---|
| Guluronic Acid | 1.20 | 1.62 |
| Mannuronic Acid | 10.25 | 9.80 |
| Mannose | 0.95 | 0.73 |
| Ribose | 0.17 | 0.00 |
| Rhamnose | 0.26 | 0.22 |
| Glucuronic Acid | 0.38 | 0.51 |
| Galacturonic Acid | 0.14 | 0.27 |
| Glucose | 3.63 | 1.89 |
| Galactose | 0.72 | 0.49 |
| Xylose | 0.56 | 0.41 |
| Arabinose | 0.25 | 0.05 |
| Fucose | 2.01 | 1.70 |
| Total | 20.52 | 17.68 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kaur, S.; Abraham, R.E.; Franco, C.M.M.; Puri, M. Production of Alginate Oligosaccharides (AOSs) Using Enhanced Physicochemical Properties of Immobilized Alginate Lyase for Industrial Application. Mar. Drugs 2024, 22, 120. https://doi.org/10.3390/md22030120
Kaur S, Abraham RE, Franco CMM, Puri M. Production of Alginate Oligosaccharides (AOSs) Using Enhanced Physicochemical Properties of Immobilized Alginate Lyase for Industrial Application. Marine Drugs. 2024; 22(3):120. https://doi.org/10.3390/md22030120
Chicago/Turabian StyleKaur, Simranjeet, Reinu E. Abraham, Christopher M. M. Franco, and Munish Puri. 2024. "Production of Alginate Oligosaccharides (AOSs) Using Enhanced Physicochemical Properties of Immobilized Alginate Lyase for Industrial Application" Marine Drugs 22, no. 3: 120. https://doi.org/10.3390/md22030120