Phylogenetic Analysis and Antimicrobial Profiles of Cultured Emerging Opportunistic Pathogens (Phyla Actinobacteria and Proteobacteria) Identified in Hot Springs
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling Sites
2.2. Isolation of Bacteria
2.3. DNA Extraction, 16S rDNA Gene Sequencing and Phylogeny
2.4. Assessment of the Presence of Legionella spp. Using Real-Time PCR
2.5. Antibiotic Resistance Assay
2.6. GenBank Accession Numbers
3. Results
3.1. Isolation Rates of Bacteria
3.2. 16S rDNA Gene Sequencing
3.3. Phylogenetic Analysis
3.4. Detection of Legionella spp.
3.5. Antibiotic Resistance
4. Discussion
4.1. Phylum Actinobacteria
4.2. Phylum Proteobacteria
4.2.1. Alphaproteobacteria
4.2.2. Betaproteobacteria
4.2.3. Gammaproteobacteria
4.3. Antibiotic Resistance of Opportunistic Emerging Pathogens
4.4. Relationship between Waterborne Pathogens and Emerging Opportunistic Pathogens
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Sharma, S.; Sachdeva, P.; Virdi, J.S. Emerging water-borne pathogens. Appl. Microbiol. Biotechnol. 2003, 61, 424–428. [Google Scholar] [CrossRef] [PubMed]
- Nel, L.; Markotter, W. Emerging infectious waterborne diseases: Bacterial agents. In Microbial Waterborne Pathogens; Cloete, T., Rose, J., Nel, L., Ford, T., Eds.; IWA Publishing: London, UK, 2004. [Google Scholar]
- Pandey, P.K.; Kass, P.H.; Soupir, M.L.; Biswas, S.; Singh, V.P. Contamination of water resources by pathogenic bacteria. AMB Express 2014, 4, 51. [Google Scholar] [CrossRef] [PubMed]
- Dewaal, C.S.; Robert, N.; Witmer, J.; Tian, X.A. A comparison of the burden of foodborne and waterborne diseases in three world regions, 2008. Food Prot. Trends 2010, 30, 483–490. [Google Scholar]
- Nasermoaddeli, A.; Kagamimori, S. Balneotherapy in medicine: A Review. Environ. Health Prev. Med. 2005, 81, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Lund, J.W. Balneological use of thermal waters. Geo-Heat Cent. Q. Bull. 1999, 20, 1–13. [Google Scholar]
- Perestrelo, M.F.; Norberg, A.N.; Guerra-Sanches, F.; Torres, A.C.; Pile, E. Microbiological analysis of water used in hydrotherapy. J. Venom. Anim. Toxins Incl. Trop. Dis. 2006, 12, 418–422. [Google Scholar] [CrossRef]
- Bhanjan, M.N.; Campus, P.; Delhi, N. Prevalence of opportunist pathogens in thermal prings of devotion. J. Appl. Sci. Environ. Sanit. 2013, 8, 195–203. [Google Scholar]
- Kurosawa, H.; Fujita, M.; Kobatake, S.; Kimura, H.; Ohshima, M.; Nagai, A.; Kaneko, S.; Iwasaki, Y.; Kozawa, K. A case of Legionella pneumonia linked to a hot spring facility in Gunma Prefecture, Japan. Jpn. J. Infect. Dis. 2010, 63, 78–79. [Google Scholar] [PubMed]
- Sukthana, Y.; Lekkla, A.; Sutthikornchai, C.; Wanapongse, P.; Vejjajiva, A. Spa, springs and safety. Southeast Asian J. Trop. Med. Public Health 2005, 36, 10–16. [Google Scholar] [PubMed]
- Fan, N.W.; Wu, C.C.; Chen, T.L.; Yu, W.K.; Chen, C.P.; Lee, S.M.; Lin, P.Y. Microsporidial keratitis in patients with hot springs exposure. J. Clin. Microbiol. 2012, 50, 414–418. [Google Scholar] [CrossRef] [PubMed]
- Yarita, K.; Sano, A.; Murata, Y.; Takayama, A.; Takahashi, Y.; Takahashi, H.; Yaguchi, T.; Ohori, A.; Kamei, K.; Miyaji, M.; et al. Pathogenicity of Ochroconis gallopava isolated from hot springs in Japan and a review of published reports. Mycopathologia 2007, 164, 135–147. [Google Scholar] [CrossRef] [PubMed]
- Drudy, D.; Mullane, N.R.; Quinn, T.; Wall, P.G.; Fanning, S. Enterobacter sakazakii: An emerging pathogen in powdered infant formula. Clin. Infect. Dis. 2006, 42, 996–1002. [Google Scholar] [CrossRef] [PubMed]
- Hunter, C.J.; Petrosyan, M.; Ford, H.R.; Prasadarao, N.V. Enterobacter sakazakii: An emerging pathogen in infants and neonates. Surg. Infect. 2008, 9, 533–539. [Google Scholar] [CrossRef] [PubMed]
- Fakruddin, M.; Rahaman, M.; Ahmed, M.M.; Hoque, M. Cronobacter sakazakii (Enterobacter sakazakii): An emerging foodborne pathogen. Int. J. Biomed. Adv. Res. 2013, 4, 350–359. [Google Scholar] [CrossRef]
- Akhtyamova, N. Human Pathogens—The plant and useful endophytes. J. Med. Microbiol. Diagnosis 2013, 2, 2–3. [Google Scholar] [CrossRef]
- Cleary, J.L.; Condren, A.R.; Zink, K.E.; Sanchez, L.M. Calling all hosts: Bacterial communication in situ. Chem 2017, 2, 334–358. [Google Scholar] [CrossRef]
- Purty, S.; Saranathan, R.; Prashanth, K.; Narayanan, K.; Asir, J.; Sheela Devi, C.; Kumar Amarnath, S. The expanding spectrum of human infections caused by Kocuria species: A case report and literature review. Emerg. Microbes Infect. 2013, 2, e71. [Google Scholar] [CrossRef] [PubMed]
- Paul, M.; Gupta, R.; Khush, S.; Thakur, R. Kocuria rosea: An emerging pathogen in acute bacterial meningitis—Case report. J. Microbiol. Antimicrob. Agents 2015, 1, 4–7. [Google Scholar]
- Sinton, L.W.; Finlay, R.K.; Lynch, P.A. Sunlight inactivation of fecal bacteriophages and bacteria in sewage-polluted seawater. Appl. Environ. Microbiol. 1999, 65, 3605–3613. [Google Scholar] [PubMed]
- Bittar, F.; Rolain, J. Detection and accurate identification of new or emerging bacteria in cystic fibrosis patients. Clin. Microbiol. Infect. 2010, 16, 809–820. [Google Scholar] [CrossRef] [PubMed]
- Van Belkum, A. Classification of bacterial pathogens. Commission on Genetic Modification (COGEM) Research Report; COGEM: Bilthoven, The Netherlands, 2011. [Google Scholar]
- Berg, G.; Eberl, L.; Hartmann, A. The rhizosphere as a reservoir for oppurtunistic human pathogenic bacteria. Environ. Microbiol. 2005, 7, 1672–1685. [Google Scholar] [CrossRef] [PubMed]
- Williams, M.M.; Armbruster, C.R.; Arduino, M.J. Plumbing of hospital premises is a reservoir for opportunistically pathogenic microorganisms: A review. Biofouling 2013, 29, 147–162. [Google Scholar] [CrossRef] [PubMed]
- Kanamori, H.; Weber, D.J.; Rutala, W.A. Healthcare outbreaks associated with a water reservoir and infection prevention strategies. Clin. Infect. Dis. 2016, 62, 1423–1435. [Google Scholar] [CrossRef] [PubMed]
- Cliver, D.O. Emerging pathogens on the rise: How can waterborne illness be prevented? Calif. Agric. 2000, 54, 78–79. [Google Scholar] [CrossRef]
- Janda, J.M.; Abbott, S.L. The genus Hafnia: From soup to nuts. Clin. Microbiol. Rev. 2006, 19, 12–28. [Google Scholar] [CrossRef] [PubMed]
- Piccini, C.; Conde, D.; Alonso, C.; Sommaruga, R.; Pernthaler, J. Blooms of single bacterial species in a coastal lagoon of the southwestern Atlantic Ocean. Appl. Environ. Microbiol. 2006, 72, 6560–6568. [Google Scholar] [CrossRef] [PubMed]
- Parthuisot, N.; West, N.J.; Lebaron, P.; Baudart, J. High diversity and abundance of Legionella spp. in a pristine river and impact of seasonal and anthropogenic effects. Appl. Environ. Microbiol. 2010, 76, 8201–8210. [Google Scholar] [CrossRef] [PubMed]
- Berthiaume, C.; Gilbert, Y.; Fournier-Larente, J.; Pluchon, C.; Filion, G.; Jubinville, E.; Sérodes, J.B.; Rodriguez, M.; Duchaine, C.; Charette, S.J. Identification of dichloroacetic acid degrading Cupriavidus bacteria in a drinking water distribution network model. J. Appl. Microbiol. 2014, 116, 208–221. [Google Scholar] [CrossRef] [PubMed]
- Vaz-Moreira, I.; Nobre, M.F.; Nunes, O.C.; Manaia, C.M. Gulbenkiania mobilis gen. nov., sp. nov., isolated from treated municipal wastewater. Int. J. Syst. Evol. Microbiol. 2007, 57, 1108–1112. [Google Scholar] [CrossRef] [PubMed]
- Khiyami, M.A.; Serour, E.A.; Shehata, M.M.; Bahklia, A.H. Thermo-aerobic bacteria from geothermal springs in Saudi Arabia. Afr. J. Biotechnol. 2012, 11, 4053–4062. [Google Scholar]
- Obeidat, M.; Khyami-Horani, H.; Otri, A.A.-Z.; Otri, I. Isolation, characterization, and hydrolytic activities of Geobacillus species from Jordanian hot springs. Afr. J. Biotechnol. 2012, 11, 6763–6768. [Google Scholar]
- Panda, M.K.; Sahu, M.K.; Tayung, K. Isolation and characterization of a thermophilic Bacillus sp. with protease activity isolated from hot spring of Tarabalo, Odisha, India. Iran. J. Microbiol. 2013, 5, 159–165. [Google Scholar] [PubMed]
- Pandey, A.; Dhakar, K.; Sharma, A.; Priti, P.; Sati, P.; Kumar, B. Thermophilic bacteria that tolerate a wide temperature and pH range colonize the Soldhar (95 °C) and Ringigad (80 °C) hot springs of Uttarakhand, India. Ann. Microbiol. 2015, 65, 809–816. [Google Scholar] [CrossRef]
- Lebedinsky, A.V.; Chernyh, N.A.; Bonch-Osmolovskaya, E.A. Phylogenetic systematics of microorganisms inhabiting thermal environments. Biochemistry 2007, 72, 1299–1312. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.K.; Lee, S.B. Isolation and characterization of a thermotolerant bacterium Ralstonia sp. Strain PHS1 that degrades benzene, toluene, ethylbenzene, and o-xylene. Appl. Microbiol. Biotechnol. 2001, 56, 270–275. [Google Scholar] [CrossRef] [PubMed]
- Berjeaud, J.M.; Chevalier, S.; Schlusselhuber, M.; Portier, E.; Loiseau, C.; Aucher, W.; Lesouhaitier, O.; Verdon, J. Legionella pneumophila: The paradox of a highly sensitive opportunistic waterborne pathogen able to persist in the environment. Front. Microbiol. 2016, 7, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Asgarani, E.; Soudi, M.R.; Borzooee, F.; Dabbagh, R. Radio-resistance in psychrotrophic Kocuria sp. ASB 107 isolated from Ab-e-Siah radioactive spring. J. Environ. Radioact. 2012, 113, 171–176. [Google Scholar] [CrossRef] [PubMed]
- Gholami, M.; Etemadifar, Z.; Bouzari, M. Isolation a new strain of Kocuria rosea capable of tolerating extreme conditions. J. Environ. Radioact. 2015, 144, 113–119. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.L.; Chou, Y.J.; Chen, W.M.; Arun, B.; Young, C.C. Tepidimonas taiwanensis sp. nov., a novel alkaline-protease-producing bacterium isolated from a hot spring. Extremophiles 2006, 10, 35–40. [Google Scholar] [CrossRef] [PubMed]
- Jaradat, Z.W.; Al Mousa, W.; Elbetieha, A.; Al Nabulsi, A.; Tall, B.D. Cronobacter spp.—Opportunistic food-borne pathogens. A review of their virulence and environmental-adaptive traits. J. Med. Microbiol. 2014, 63, 1023–1037. [Google Scholar] [CrossRef] [PubMed]
- Joint United Nations Programme on HIV/AIDS (UNAIDS). Ending AIDS: Progress towards the 90-90-90 Targets. Global AIDS Update 2017; UNAIDS: Geneva, Switzerland, 2017. [Google Scholar]
- Statistics South Africa. Statistical Release P0302. Mid-Year Population Estimate 2016; Statistics South Africa: Pretoria, South Africa, 2016.
- Said-Mohamed, R.; Micklesfield, L.K.; Pettifor, J.M.; Norris, S.A. Has the prevalence of stunting in South African children changed in 40 years? A systematic review. BMC Public Health 2015, 15, 534. [Google Scholar] [CrossRef] [PubMed]
- Dashti, A.A.; Jadaon, M.M.; Abdulsamad, A.M.; Dashti, H.M. Heat treatment of bacteria: A simple method of DNA extraction for molecular techniques. Kuwait Med. J. 2009, 41, 117–122. [Google Scholar]
- Olivier, J.; Venter, J.S.; Jonker, C. Thermal and chemical characteristics of hot water springs in the northern part of the Limpopo Province, South Africa. Water SA 2011, 37, 427–436. [Google Scholar] [CrossRef]
- Galkiewicz, J.P.; Kellogg, C.A. Cross-kingdom amplification using Bacteria-specific primers: Complications for studies of coral microbial ecology. Appl. Environ. Microbiol. 2008, 74, 7828–7831. [Google Scholar] [CrossRef] [PubMed]
- McGinnis, S.; Madden, T.L. BLAST: At the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 2004, 32, 20–25. [Google Scholar] [CrossRef] [PubMed]
- Kim, O.S.; Cho, Y.J.; Lee, K.; Yoon, S.H.; Kim, M.; Na, H.; Park, S.C.; Jeon, Y.S.; Lee, J.H.; Yi, H.; et al. Introducing EzTaxon-e: A prokaryotic 16s rRNA gene sequence database with phylotypes that represent uncultured species. Int. J. Syst. Evol. Microbiol. 2012, 62, 716–721. [Google Scholar] [CrossRef] [PubMed]
- Yarza, P.; Yilmaz, P.; Pruesse, E.; Glöckner, F.O.; Ludwig, W.; Schleifer, K.-H.; Whitman, W.B.; Euzéby, J.; Amann, R.; Rosselló-Móra, R. Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat. Rev. Microbiol. 2014, 12, 635–645. [Google Scholar] [CrossRef] [PubMed]
- Gouy, M.; Guindon, S.; Gascuel, O. SeaView Version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 2010, 27, 221–224. [Google Scholar] [CrossRef] [PubMed]
- Wellinghausen, N.; Frost, C.; Marre, R. Detection of Legionellae in hospital water samples by quantitative real-time LightCycler PCR. Appl. Environ. Microbiol. 2001, 67, 3985–3993. [Google Scholar] [CrossRef] [PubMed]
- Bauer, A.W.; Kirby, W.M.; Sherris, J.C.; Turck, M. Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Pathol. 1966, 45, 493–496. [Google Scholar] [PubMed]
- Clinical and Laboratory Standards Institute (CLSI) M02-A12. Performance Standards for Antimicrobial Disk Susceptibility Tests; Approved Standard—Twelfth Edition; CLSI: Wayne, NJ, USA, 2015. [Google Scholar]
- Middleton, J.H.; Ambrose, A. Enumeration and antibiotic resistance patterns of fecal indicator organisms isolated from migratory Canada geese (Branta canadensis). J. Wildl. Dis. 2005, 41, 334–341. [Google Scholar] [CrossRef] [PubMed]
- Yakhlef, W.; Darbouche, A. Metabolic diversity of thermophilic bacteria from hot springs in Algeria. J. Acad. 2012, 2, 57–65. [Google Scholar]
- Thorolfsdottir, B.; Marteinsson, V. Microbiological analysis in three diverse natural geothermal bathing pools in Iceland. Int. J. Environ. Res. Public Health 2013, 10, 1085–1099. [Google Scholar] [CrossRef] [PubMed]
- Zhou, G.; Luo, X.; Tang, Y.; Zhang, L.; Yang, Q.; Qiu, Y.; Fang, C.X. Kocuria flava sp. nov. and Kocuria turfanensis sp. nov., airborne actinobacteria isolated from Xinjiang, China. Int. J. Syst. Evol. Microbiol. 2008, 58, 1304–1307. [Google Scholar] [CrossRef] [PubMed]
- Savini, V.; Catavitello, C.; Masciarelli, G.; Astolfi, D.; Balbinot, A.; Bianco, A.; Febbo, F.; D’Amario, C.; D’Antonio, D. Drug sensitivity and clinical impact of members of the genus Kocuria. J. Med. Microbiol. 2010, 59, 1395–1402. [Google Scholar] [CrossRef] [PubMed]
- Ma, E.S.; Wong, C.L.; Lai, K.T.; Chan, E.C.; Yam, W.; Chan, A.C. Kocuria kristinae infection associated with acute cholecystitis. BMC Infect. Dis. 2005, 5, 60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, H.; Jogler, M.; Tindall, B.J.; Klenk, H.P.; Rohde, M.; Busse, H.J.; Overmann, J. Sphingomonas starnbergensis sp. nov., isolated from a prealpine freshwater lake. Int. J. Syst. Evol. Microbiol. 2013, 63, 1017–1023. [Google Scholar] [CrossRef] [PubMed]
- Bhaiyat, M.; Hariharan, H.; Chikweto, A.; Tiwari, K.; Sharma, R.; Kobayashi, Y. Isolation of coagulase-negative Staphylococcus spp. and Kocuria varians in pure culture from tissues of cases of mortalities in parrots in Grenada, West Indies. Int. J. Vet. Med. Res. Rep. 2013, 2013, 1–6. [Google Scholar] [CrossRef]
- Yoshinaka, T.; Yano, K.; Yamaguchi, H. Isolation of highly radioresistant bacterium, Arthrobacter radiotolerans nov. sp. Agric. Biol. Chem. 1973, 37, 2269–2275. [Google Scholar] [CrossRef]
- Yang, J.; Li, X.; Huang, L.; Jiang, H. Actinobacterial diversity in the sediments of five cold springs on the Qinghai-Tibet Plateau. Front. Microbiol. 2015, 6, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Janssens, L.E.; Wauters, G.; Charlier, J.; Delmee, M. Identification of Arthrobacter oxydans, Arthrobacter luteolus sp. nov., and Arthrobacter albus sp. nov., isolated from human clinical specimens. J. Clin. Microbiol. 2000, 38, 2412–2415. [Google Scholar]
- Zhou, L.; Li, H.; Zhang, Y.; Han, S.; Xu, H. Development of genus-specific primers for better understanding the diversity and population structure of Sphingomonas in soils. J. Basic Microbiol. 2014, 54, 880–888. [Google Scholar] [CrossRef] [PubMed]
- Briggs, B.R.; Brodie, E.L.; Tom, L.M.; Dong, H.; Jiang, H.; Huang, Q.; Wang, S.; Hou, W.; Wu, G.; Huang, L.; et al. Seasonal patterns in microbial communities inhabiting the hot springs of Tengchong, Yunnan Province, China. Environ. Microbiol. 2014, 16, 1579–1591. [Google Scholar] [CrossRef] [PubMed]
- Coenye, T.; Vandamme, P.; Lipuma, J.J. Infection by Ralstonia species in cystic fibrosis patients: Identification of R. pickettii and R. mannitolilytica by polymerase chain reaction. Emerg. Infect. Dis. 2002, 8, 692–696. [Google Scholar] [CrossRef] [PubMed]
- Langevin, S.; Vincelette, J.; Bekal, S.; Gaudreau, C. First case of invasive human infection caused by Cupriavidus metallidurans. J. Clin. Microbiol. 2011, 49, 744–745. [Google Scholar] [CrossRef] [PubMed]
- Karafin, M.; Romagnoli, M.; Fink, D.L.; Howard, T.; Rau, R.; Milstone, A.M.; Carroll, K.C. Fatal infection caused by Cupriavidus gilardii in a child with aplastic anemia. J. Clin. Microbiol. 2010, 48, 1005–1007. [Google Scholar] [CrossRef] [PubMed]
- Ko, K.S.; Lee, N.Y.; Oh, W.S.; Lee, J.H.; Ki, H.K.; Peck, K.R.; Song, J.H. Tepidimonas arfidensis sp. Nov., a novel Gram-negative and thermophilic bacterium isolated from the bone marrow of a patient with leukemia in Korea. Microbiol. Immunol. 2005, 49, 785–788. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.M.; Huang, H.W.; Chang, J.S.; Han, Y.L.; Guo, T.R.; Sheu, S.Y. Tepidimonas fonticaldi sp. nov., a slightly thermophilic betaproteobacterium isolated from a hot spring. Int. J. Syst. Evol. Microbiol. 2013, 63, 1810–1816. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Liu, W.; Cui, L.; Zhang, M.; Wang, B. Characterization and identification of a chlorine-resistant bacterium, Sphingomonas TS001, from a model drinking water distribution system. Sci. Total Environ. 2013, 458–460, 169–175. [Google Scholar] [CrossRef] [PubMed]
- Jimat, D.N.; Baizura, I.; Mohamed, F.; Azmi, A.S.; Zainudin, Z. Isolation and characterization of thermophilic bacteria producing L-Asparaginase from Malaysia hotspring and enzyme activity using different carbon and nitrogen sources. J. Appl. Sci. Agric. 2015, 10, 69–77. [Google Scholar]
- Huys, G.; Cnockaert, M.; Abbott, S.L.; Janda, J.M.; Vandamme, P. Hafnia paralvei sp. nov., formerly known as Hafnia alvei hybridization group 2. Int. J. Syst. Evol. Microbiol. 2010, 60, 1725–1728. [Google Scholar] [CrossRef] [PubMed]
- Qin, T.; Yan, G.; Ren, H.; Zhou, H.; Wang, H.; Xu, Y.; Zhao, M.; Guan, H.; Li, M.; Shao, Z. High prevalence, genetic diversity and intracellular growth ability of Legionella in hot spring environments. PLoS ONE 2013, 8, e59018. [Google Scholar] [CrossRef] [PubMed]
- Walczak, M.; Burkowska-But, A.; Swiontek Brzezinska, M.; Krawiec, A. Distribution of Legionella pneumophila in thermal pools. CLEAN 2016, 44, 532–537. [Google Scholar]
- Kobayashi, M.; Oana, K.; Kawakami, Y. Bath water contamination with Legionella and nontuberculous mycobacteria in 24-hour home baths, hot springs, and public bathhouses of Nagano Prefecture, Japan. Jpn. J. Infect. Dis. 2014, 67, 276–281. [Google Scholar] [CrossRef] [PubMed]
- Kao, P.M.; Tung, M.C.; Hsu, B.M.; Hsueh, C.J.; Chiu, Y.C.; Chen, N.H.; Shen, S.M.; Huang, Y.L. Occurrence and distribution of Naegleria species from thermal spring environments in Taiwan. Lett. Appl. Microbiol. 2013, 56, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Nour, M.; Duncan, C.; Low, D.E.; Guyard, C. Biofilms: The stronghold of Legionella pneumophila. Int. J. Mol. Sci. 2013, 14, 21660–21675. [Google Scholar] [CrossRef] [PubMed]
- Abbott, S.L.; Moler, S.; Green, N.; Tran, R.K.; Wainwright, K.; Janda, J.M. Clinical and laboratory diagnostic characteristics and cytotoxigenic potential of Hafnia alvei and Hafnia paralvei strains. J. Clin. Microbiol. 2011, 49, 3122–3126. [Google Scholar] [CrossRef] [PubMed]
- Brandão, M.L.L.; Umeda, N.S.; Jackson, E.; Forsythe, S.J.; de Filippis, I. Isolation, molecular and phenotypic characterization, and antibiotic susceptibility of Cronobacter spp. from Brazilian retail foods. Food Microbiol. 2017, 63, 129–138. [Google Scholar] [CrossRef] [PubMed]
- Kilonzo-Nthenge, A.; Rotich, E.; Godwin, S.; Nahashon, S.; Chen, F. Prevalence and antimicrobial resistance of Cronobacter sakazakii isolated from domestic kitchens in middle Tennessee, United States. J. Food Prot. 2012, 75, 1512–1517. [Google Scholar] [CrossRef] [PubMed]
- Maartens, G. Opportunistic infections associated with HIV infection in Africa. Oral Dis 2002, 8 (Suppl. 2), 76–79. [Google Scholar] [CrossRef] [PubMed]
- Holmes, C.B.; Losina, E.; Walensky, R.P.; Yazdanpanah, Y.; Freedberg, K.A. Review of human immunodeficiency virus type 1-related opportunistic infections in sub-Saharan Africa. Clin. Infect. Dis. 2003, 36, 652–662. [Google Scholar] [CrossRef] [PubMed]
- Marais, B.J.; Rabie, H.; Schaaf, S.H.; Cotton, M.F. Common opportunistic infections in HIV infected infants and children Part 1—Respiratory infections. S. Afri. Fam. Pract. 2006, 48, 1–4. [Google Scholar] [CrossRef]
- Coman, I.; Bilodeau, L.; Lavoie, A.; Carricart, M.; Tremblay, F.; Zlosnik, J.E.; Berthiaume, Y. Ralstonia mannitolilytica in cystic fibrosis: A new predictor of worse outcomes. Respir. Med. Case Rep. 2017, 20, 48–50. [Google Scholar] [CrossRef] [PubMed]
- Dunn, R.; Bares, S.; David, M.Z. Central venous catheter-related bacteremia caused by Kocuria kristinae: Case report and review of the literature. Ann. Clin. Microbiol. Antimicrob. 2011, 10, 31. [Google Scholar] [CrossRef] [PubMed]
- Al-Grawi, J.G.A. Hafnia alvei urinary tract infection. Iraqi Postgrad. Med. J. 2008, 7, 71–76. [Google Scholar]
- Lai, K.K. Enterobacter sakazakii infections among neonates, infants, children, and adults: Case reports and a review of the literature. Medicine 2001, 80, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Toh, H.S.; Tay, H.T.; Kuar, W.K.; Weng, T.C.; Tang, H.J.; Tan, C.K. Risk factors associated with Sphingomonas paucimobilis infection. J. Microbiol. Immunol. Infect. 2011, 44, 289–295. [Google Scholar] [CrossRef] [PubMed]
- Lewis, D.A.; Gumede, L.Y.E.; Van der Hoven, L.A.; De Gita, G.D.; De Kock, E.J.E.; De Lange, T.; Maseko, V.; Kekana, V.; Smuts, F.P.; Perovic, O. Antimicrobial susceptibility of organisms causing community-acquired urinary tract infections in Gauteng Province, South Africa. S. Afr. Med. J. 2013, 103, 377–381. [Google Scholar] [CrossRef] [PubMed]
- Dewan, E.; Oberoi, A.; Mathias, A. An emerging threat: A rare non-fermenter in an immunocompromised patient—Sphingomonas paucimobilis from a tertiary care centre. Indian J. Med. Case Rep. 2014, 3, 35–39. [Google Scholar]
- Ryan, M.P.; Adley, C.C. Ralstonia spp.: Emerging global opportunistic pathogens. Eur. J. Clin. Microbiol. Infect. Dis. 2014, 33, 291–304. [Google Scholar] [CrossRef] [PubMed]
- Meric, M.; Willke, A.; Kolayli, F.; Yavuz, S.; Vahaboglu, H. Water-borne Sphingomonas paucimobilis epidemic in an intensive care unit. J. Infect. 2009, 58, 253–255. [Google Scholar] [CrossRef] [PubMed]
- Coenye, T.; Goris, J.; De Vos, P.; Vandamme, P.; LiPuma, J.J. Classification of Ralstonia pickettii-like isolates from the environment and clinical samples as Ralstonia insidiosa sp. nov. Int. J. Syst. Evol. Microbiol. 2003, 53, 1075–1080. [Google Scholar] [CrossRef] [PubMed]
- Memory, T.; Prinsloo, A.; Olivier, J.; Jonker, N.; Venter, S. An evaluation of the bacterial diversity at Tshipise, Mphephu and Sagole hot water springs, Limpopo Province, South Africa. Afr. J. Microbiol. Res. 2012, 6, 4993–5004. [Google Scholar]
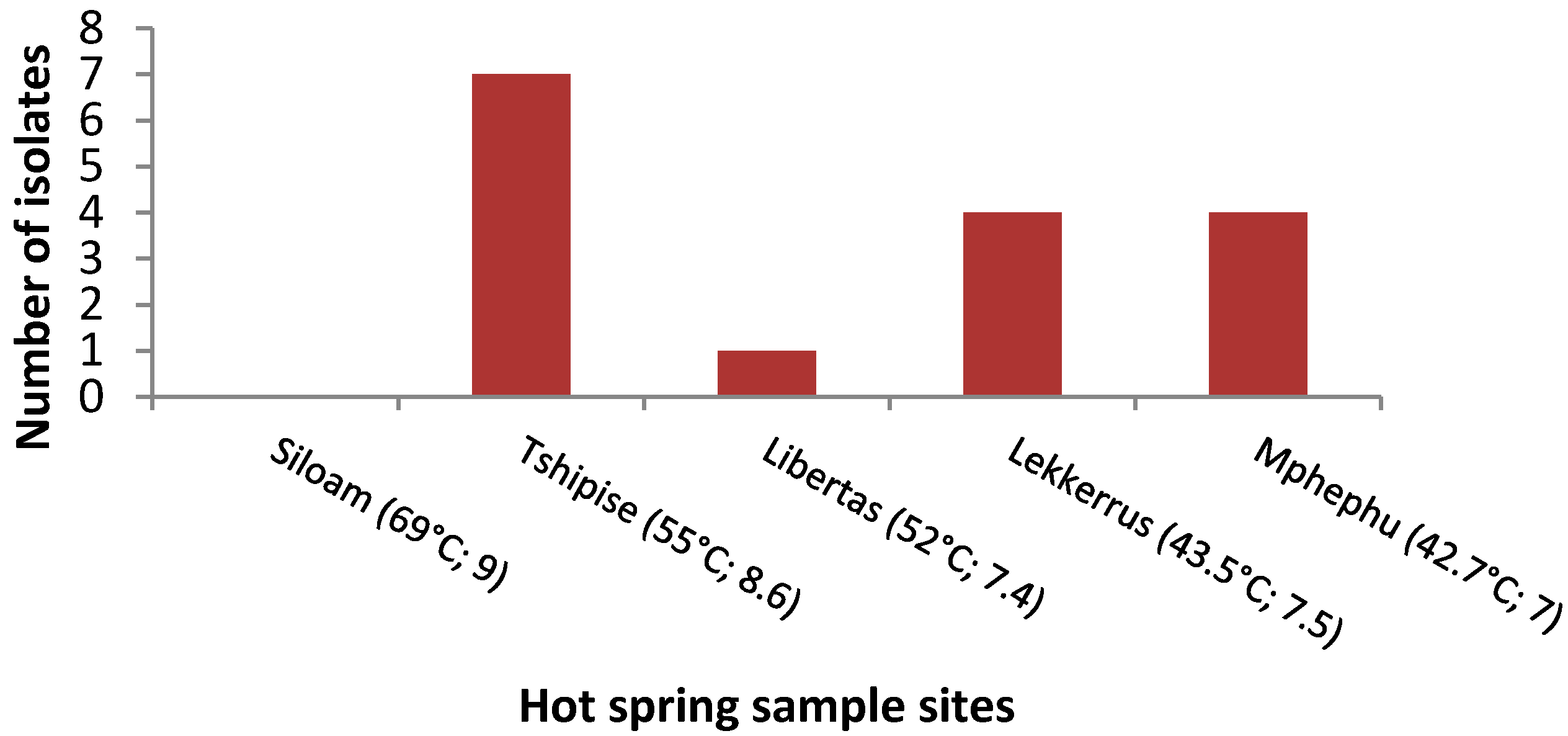
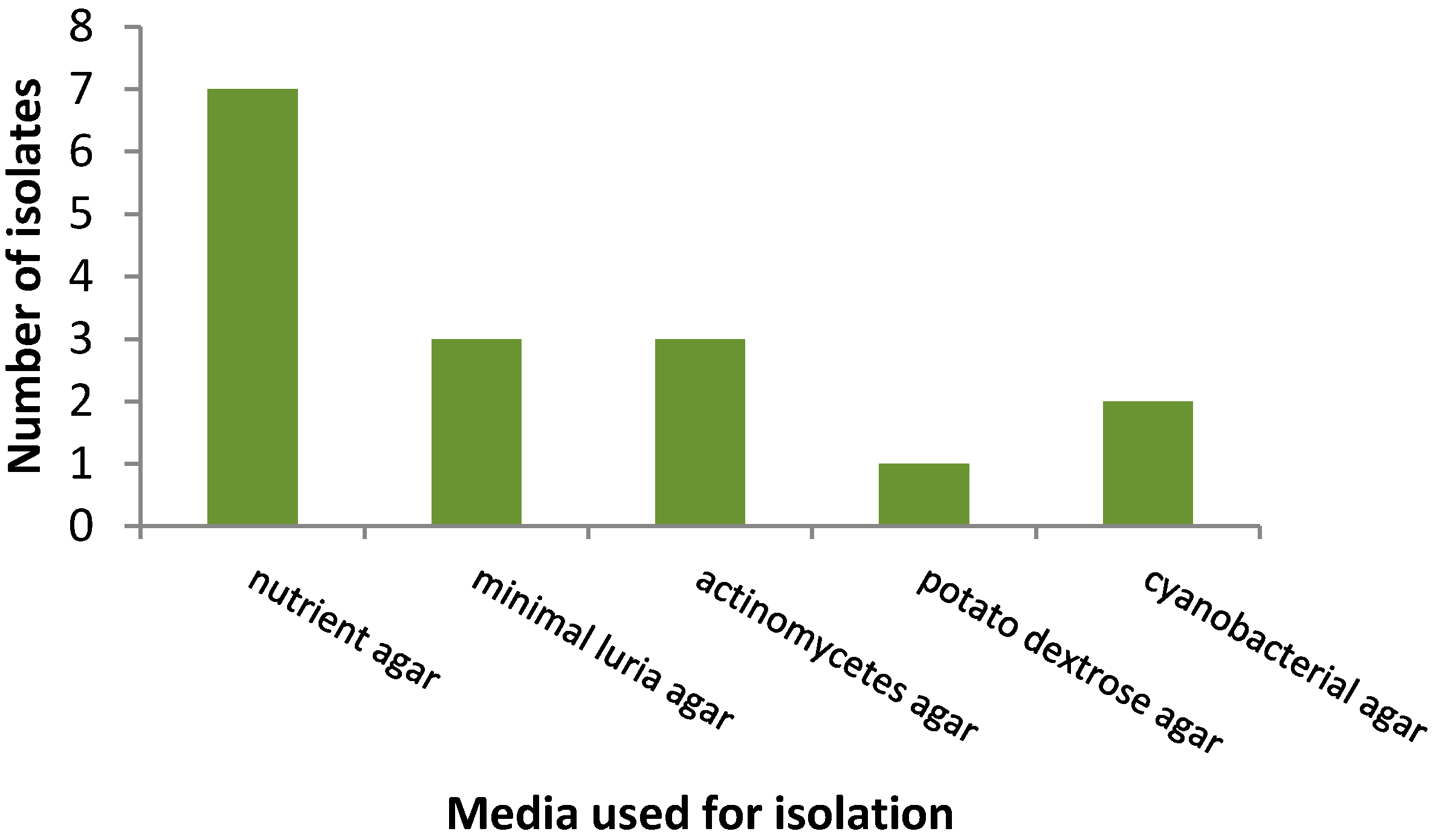
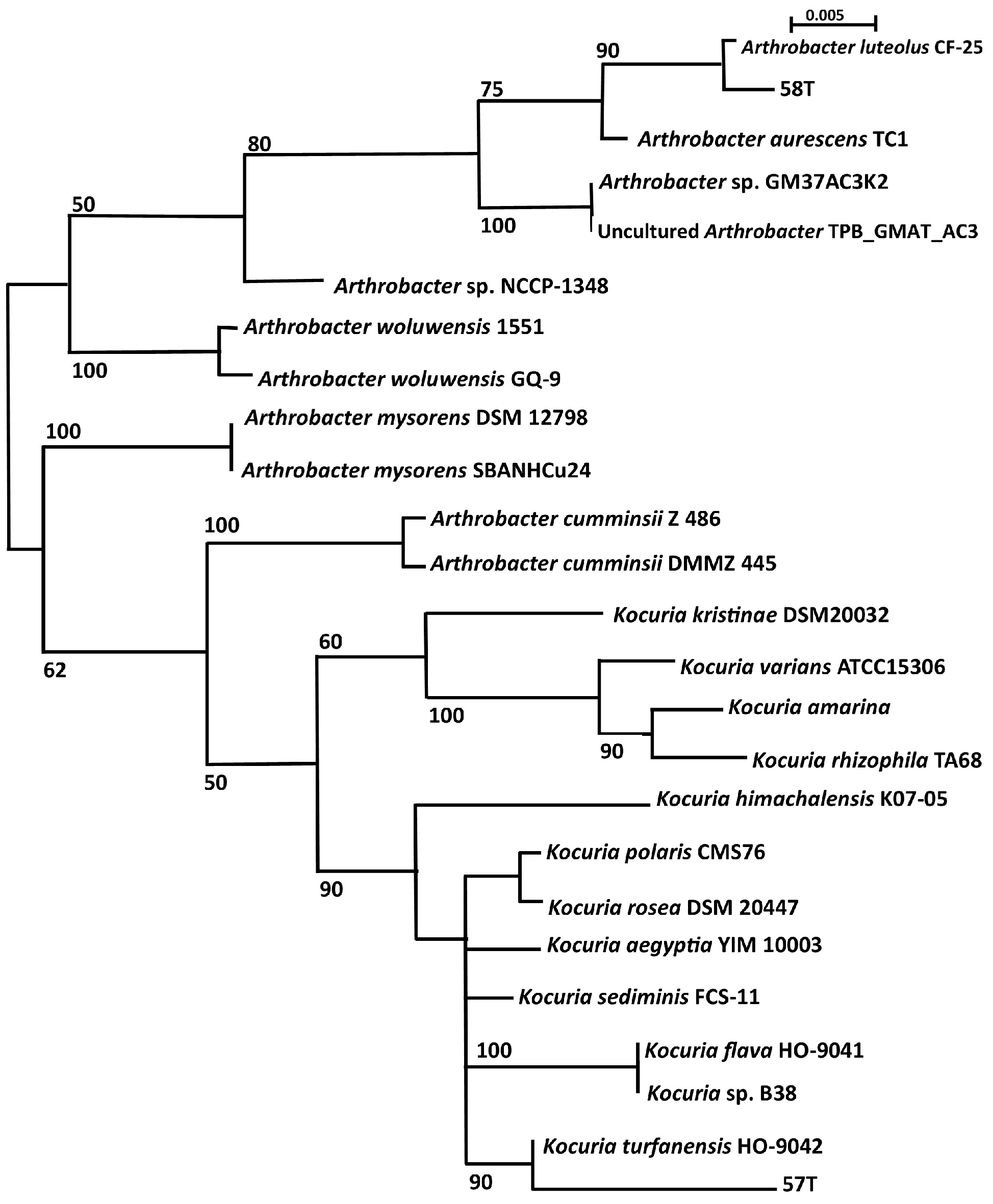
| Sampling Site | GPS Location | pH | Temperature (°C) | Comments |
|---|---|---|---|---|
| Tshipise | 22°36.521′ S 30°10.345′ E | 8.63 | 55.2 | Open to air in enclosed section |
| Siloam | 22°53.667′ S 30° 11.7718′ E | 9 | 69 | Exiting from pipe on private property |
| Mphephu | 22°54.225′ S 30° 10.83′ E | 7.07 | 42.4 | Open to air in enclosed section |
| Lekkerrus | 24°28.04′ S 28°33.1′ E | 7.46 | 43.5 | Pipeline conveys water into pool; flow manually controlled |
| Libertas | 24°27′36″ S 28°34′11″ E | 7.44 | 52.1 | Water pumped at source |
| Isolate No. | Site | Isolation Temperature (°C) | Sample | Isolation Media |
|---|---|---|---|---|
| 57T | Tshipise | 37 | Water | Nutrient agar |
| 58T | Tshipise | 37 | Water | Actinomycete isolation agar |
| 87T | Tshipise | 25 | Water | Minimal Luria agar |
| 61T | Tshipise | 25 | Water | Cyanobacterial agar |
| 72T | Tshipise | 37 | Water | Nutrient agar |
| 80Lk | Lekkerrus | 37 | Water | Nutrient agar |
| 79M | Mphephu | 37 | Water | Nutrient agar |
| 44M | Mphephu | 53 | Sediment | Nutrient agar |
| 55M | Mphephu | 37 | Water | Potato dextrose agar |
| 37Lb | Libertas | 53 | Water | Actinomycete isolation agar |
| 42T | Tshipise | 53 | Sediment | Nutrient agar |
| 59Lk | Lekkerrus | 37 | Water | Minimal Luria agar |
| 5T | Tshipise | 53 | Water | Minimal Luria agar |
| 27M | Mphephu | 53 | Water | Actinomycete isolation agar |
| 69Lk | Lekkerrus | 25 | Water | Cyanobacterial agar |
| 31Lk | Lekkerrus | 53 | Water | Nutrient agar |
| Isolate No. | Identification | CAR100 | GEN10 | KAN30 | STR10 | TET30 | CEF30 | CHL30 | COT25 | NA30 | NOR10 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 57T | Kocuria turfanensis | 15 | 9 | 12 | 7 | 12 | 0 | 14 | 16 | 0 | 2 |
| 58T | Arthrobacter luteolus | 0 | 5 | 3 | 4 | 9 | 1 | 6 | 11 | 0 | 2 |
| 79M | Hafnia alvei | 0 | 2 | 3 | 1 | 4 | 0 | 5 | 6 | 3 | 7 |
| 80Lk | Cronobacter sp. | 1 | 4 | 5 | 5 | 6 | 2 | 6 | 6 | 3 | 11 |
| 72T | unknown Enterobacteriaceae | 5 | 6 | 10 | 5 | 7 | 0 | 0 | 16 | 5 | 14 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jardine, J.L.; Abia, A.L.K.; Mavumengwana, V.; Ubomba-Jaswa, E. Phylogenetic Analysis and Antimicrobial Profiles of Cultured Emerging Opportunistic Pathogens (Phyla Actinobacteria and Proteobacteria) Identified in Hot Springs. Int. J. Environ. Res. Public Health 2017, 14, 1070. https://doi.org/10.3390/ijerph14091070
Jardine JL, Abia ALK, Mavumengwana V, Ubomba-Jaswa E. Phylogenetic Analysis and Antimicrobial Profiles of Cultured Emerging Opportunistic Pathogens (Phyla Actinobacteria and Proteobacteria) Identified in Hot Springs. International Journal of Environmental Research and Public Health. 2017; 14(9):1070. https://doi.org/10.3390/ijerph14091070
Chicago/Turabian StyleJardine, Jocelyn Leonie, Akebe Luther King Abia, Vuyo Mavumengwana, and Eunice Ubomba-Jaswa. 2017. "Phylogenetic Analysis and Antimicrobial Profiles of Cultured Emerging Opportunistic Pathogens (Phyla Actinobacteria and Proteobacteria) Identified in Hot Springs" International Journal of Environmental Research and Public Health 14, no. 9: 1070. https://doi.org/10.3390/ijerph14091070






