Characterization and Dynamic Shift of Microbial Communities during Start-Up, Overloading and Steady-State in an Anaerobic Membrane Bioreactor
Abstract
:1. Introduction
2. Materials and Methods
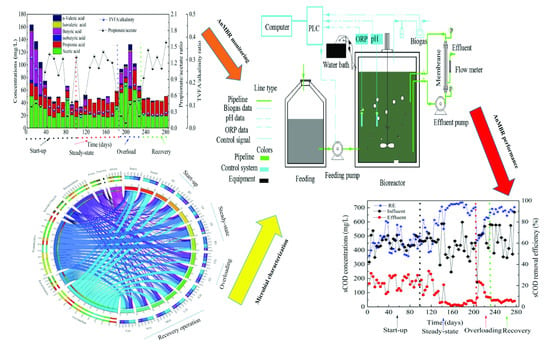
2.1. Anaerobic Membrane Bioreactor Operation
2.2. Inoculum, Sample Collection and Operational Parameters of the AnMBR
2.3. Microbial Community Analysis
2.4. Analytical Methods
3. Results
3.1. Performance of the AnMBR Bioreactor
3.2. Microbial Community Evolution
3.2.1. Microbial Community Composition and Shift
3.2.2. Archaea and Bacteria Dynamic Populations at the Genus and Family
3.2.3. Similarity, Richness and Diversity among AnMBR Microbial Community
4. Discussion
Microbial Community Shift and Contribution in the AnMBR
5. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Bandara, W.M.; Kindaichi, T.; Satoh, H.; Sasakawa, M.; Nakahara, Y.; Takahashi, M.; Okabe, S. Anaerobic treatment of municipal wastewater at ambient temperature: Analysis of archaeal community structure and recovery of dissolved methane. Water Res. 2012, 46, 756–5764. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Sosa, D.; Helmreich, B.; Netter, T.; Paris, S.; Bischof, F.; Horn, H. Anaerobic submerged membrane bioreactor (AnSMBR) for municipal wastewater treatment under mesophilic and psychrophilic temperature conditions. Bioresour. Technol. 2011, 102, 10377–10385. [Google Scholar] [CrossRef] [PubMed]
- Zuo, Z.; Wu, S.; Zhang, W.; Dong, R. Effects of organic loading rate and effluent recirculation on the performance of two-stage anaerobic digestion of vegetable waste. Bioresour. Technol. 2013, 146, 556–561. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Cheng, J.J.; Creamer, K.S. Inhibition of anaerobic digestion process: A review. Bioresour. Technol. 2008, 99, 4044–4064. [Google Scholar] [CrossRef] [PubMed]
- De Vrieze, J.; Verstraete, W.; Boon, N. Repeated pulse feeding induces functional stability in anaerobic digestion. Microb. Biotechnol. 2013, 6, 414–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Jonge, N.; Moset, V.; Møller, H.B.; Nielsen, J.L. Microbial population dynamics in continuous anaerobic digester systems during start up, stable conditions and recovery after starvation. Bioresour. Technol. 2017, 232, 313–320. [Google Scholar] [CrossRef] [PubMed]
- McMahon, K.D.; Stroot, P.G.; Mackie, R.I.; Raskin, L. Anaerobic codigestion of municipal solid waste and biosolids under various mixing conditions-II: Microbial population dynamics. Water Res. 2001, 35, 1817–1827. [Google Scholar] [CrossRef]
- Barber, W.P.; Stuckey, D.C. The use of the anaerobic baffled reactor (ABR) for wastewater treatment: A review. Water Res. 1999, 33, 1559–1578. [Google Scholar] [CrossRef]
- Pretel, R.; Robles, A.; Ruano, M.; Seco, A.; Ferrer, J. Economic and environmental sustainability of submerged anaerobic MBR-based (AnMBR-based) technology as compared to aerobic-based technologies for moderate-/high-loaded urban wastewater treatment. J. Environ. Manag. 2016, 166, 45–54. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; He, Q.; Ma, Y.; Wang, X.; Peng, X. Dynamics of microbial community in a mesophilic anaerobic digester treating food waste: Relationship between community structure and process stability. Bioresour. Technol. 2015, 189, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Ho, J.; Sung, S. Methanogenic activities in anaerobic membrane bioreactors (AnMBR) treating synthetic municipal wastewater. Bioresour. Technol. 2010, 101, 2191–2196. [Google Scholar] [CrossRef] [PubMed]
- Gulhane, M.; Pandit, P.; Khardenavis, A.; Singh, D.; Purohit, H. Study of microbial community plasticity for anaerobic digestion of vegetable waste in Anaerobic Baffled Reactor. Renew. Energy 2017, 101, 59–66. [Google Scholar] [CrossRef]
- Luo, G.; Li, J.; Li, Y.; Wang, Z.; Li, W.-T.; Li, A.-M. Performance, kinetics behaviors and microbial community of internal circulation anaerobic reactor treating wastewater with high organic loading rate: Role of external hydraulic circulation. Bioresour. Technol. 2016, 222, 470–477. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, A.S.; Hashsham, S.A.; Dollhopf, S.L.; Raskin, L.; Glagoleva, O.; Dazzo, F.B.; Hickey, R.F.; Criddle, C.S.; Tiedje, J.M. Flexible community structure correlates with stable community function in methanogenic bioreactor communities perturbed by glucose. Appl. Environ. Microbiol. 2000, 66, 4058–4067. [Google Scholar] [CrossRef] [PubMed]
- Steinberg, L.M.; Regan, J.M. Response of lab-scale methanogenic reactors inoculated from different sources to organic loading rate shocks. Bioresour. Technol. 2011, 102, 8790–8798. [Google Scholar] [CrossRef] [PubMed]
- Martin Vincent, N.; Tong, J.; Yu, D.; Zhang, J.; Wei, Y. Membrane Fouling Characteristics of a Side-Stream Tubular Anaerobic Membrane Bioreactor (AnMBR) Treating Domestic Wastewater. Processes 2018, 6, 50. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Federation, W.E.; American Public Health Association. Standard Methods for the Examination of Water and Wastewater; American Public Health Association (APHA): Washington, DC, USA, 2005. [Google Scholar]
- Inanc, B.; Matsui, S.; Ide, S. Propionic acid accumulation in anaerobic digestion of carbohydrates: An investigation on the role of hydrogen gas. Water Sci. Technol. 1999, 40, 93–100. [Google Scholar] [CrossRef]
- Lens, P.; Visser, A.; Janssen, A.; Pol, L.H.; Lettinga, G. Biotechnological treatment of sulfate-rich wastewaters. Crit. Rev. Environ. Sci. Technol. 1998, 28, 41–88. [Google Scholar] [CrossRef]
- Town, J.R.; Links, M.G.; Fonstad, T.A.; Dumonceaux, T.J. Molecular characterization of anaerobic digester microbial communities identifies microorganisms that correlate to reactor performance. Bioresour. Technol. 2014, 151, 249–257. [Google Scholar] [CrossRef] [PubMed]
- Cardinali-Rezende, J.; Colturato, L.F.; Colturato, T.D.; Chartone-Souza, E.; Nascimento, A.M.; Sanz, J.L. Prokaryotic diversity and dynamics in a full-scale municipal solid waste anaerobic reactor from start-up to steady-state conditions. Bioresour. Technol. 2012, 119, 373–383. [Google Scholar] [CrossRef] [PubMed]
- Regueiro, L.; Lema, J.M.; Carballa, M. Key microbial communities steering the functioning of anaerobic digesters during hydraulic and organic overloading shocks. Bioresour. Technol. 2015, 197, 208–216. [Google Scholar] [CrossRef] [PubMed]
- Lerm, S.; Kleyböcker, A.; Miethling-Graff, R.; Alawi, M.; Kasina, M.; Liebrich, M.; Würdemann, H. Archaeal community composition affects the function of anaerobic co-digesters in response to organic overload. Waste Manag. 2012, 32, 389–399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boe, K.; Batstone, D.J.; Steyer, J.-P.; Angelidaki, I. State indicators for monitoring the anaerobic digestion process. Water Res. 2010, 44, 5973–5980. [Google Scholar] [CrossRef] [PubMed]
- Ahring, B.K.; Sandberg, M.; Angelidaki, I. Volatile fatty acids as indicators of process imbalance in anaerobic digestors. Appl. Microbiol. Biotechnol. 1995, 43, 559–565. [Google Scholar] [CrossRef]
- Pullammanappallil, P.C.; Chynoweth, D.P.; Lyberatos, G.; Svoronos, S.A. Stable performance of anaerobic digestion in the presence of a high concentration of propionic acid. Bioresour. Technol. 2001, 78, 165–169. [Google Scholar] [CrossRef]
- Kleyböcker, A.; Liebrich, M.; Verstraete, W.; Kraume, M.; Würdemann, H. Early warning indicators for process failure due to organic overloading by rapeseed oil in one-stage continuously stirred tank reactor, sewage sludge and waste digesters. Bioresour. Technol. 2012, 123, 534–541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Werner, J.J.; Garcia, M.L.; Perkins, S.D.; Yarasheski, K.E.; Smith, S.R.; Muegge, B.D.; Stadermann, F.J.; DeRito, C.M.; Floss, C.; Madsen, E.L. Microbial community dynamics and stability during an ammonia-induced shift to syntrophic acetate oxidation. Appl. Environ. Microbiol. 2014, 80, 3375–3383. [Google Scholar] [CrossRef] [PubMed]
- Cho, K.; Lee, J.; Kim, W.; Hwang, S. Behavior of methanogens during start-up of farm-scale anaerobic digester treating swine wastewater. Process Biochem. 2013, 48, 1441–1445. [Google Scholar] [CrossRef]
- Wang, Z.; Yang, S.-T. Propionic acid production in glycerol/glucose co-fermentation by Propionibacterium freudenreichii subsp. shermanii. Bioresour. Technol. 2013, 137, 116–123. [Google Scholar] [CrossRef] [PubMed]
- De Vrieze, J.; Saunders, A.M.; He, Y.; Fang, J.; Nielsen, P.H.; Verstraete, W.; Boon, N. Ammonia and temperature determine potential clustering in the anaerobic digestion microbiome. Water Res. 2015, 75, 312–323. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Li, H.; Wang, G.; Wang, X. Effects of loading rate and temperature on anaerobic co-digestion of food waste and waste activated sludge in a high frequency feeding system, looking in particular at stability and efficiency. Bioresour. Technol. 2017, 237, 231–239. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Chen, X.; Liu, Z.; Zhou, X.; Zhang, Y. Effect of inoculum sources on the anaerobic digestion of rice straw. Bioresour. Technol. 2014, 158, 149–155. [Google Scholar] [CrossRef] [PubMed]
- Quintero, M.; Castro, L.; Ortiz, C.; Guzmán, C.; Escalante, H. Enhancement of starting up anaerobic digestion of lignocellulosic substrate: Fique’s bagasse as an example. Bioresour. Technol. 2012, 108, 8–13. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Chen, X.; Dai, R.; Xiang, X.; Li, G.; Cao, J.; Xue, Z.; Shang, K. Performance and Bacterial Community Change during the Start-Up Period of a Novel Anaerobic Bioreactor Inoculated with Long-Time Storage Anaerobic Granular Sludge. J. Environ. Eng. 2017, 143, 04017036. [Google Scholar] [CrossRef]
- Supaphol, S.; Jenkins, S.N.; Intomo, P.; Waite, I.S.; O’Donnell, A.G. Microbial community dynamics in mesophilic anaerobic co-digestion of mixed waste. Bioresour. Technol. 2011, 102, 4021–4027. [Google Scholar] [CrossRef] [PubMed]
- Cerrillo, M.; Morey, L.; Viñas, M.; Bonmatí, A. Assessment of active methanogenic archaea in a methanol-fed upflow anaerobic sludge blanket reactor. Appl. Microbiol. Biotechnol. 2016, 100, 10137–10146. [Google Scholar] [CrossRef] [PubMed]
- Karakashev, D.; Batstone, D.J.; Trably, E.; Angelidaki, I. Acetate oxidation is the dominant methanogenic pathway from acetate in the absence of Methanosaetaceae. Appl. Environ. Microbiol. 2006, 72, 5138–5141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oren, A. The Family Methanotrichaceae. In The Prokaryotes; Springer: Berlin, Germany, 2014; pp. 297–306. [Google Scholar]
- Honda, T.; Fujita, T.; Tonouchi, A. Aminivibrio pyruvatiphilus gen. nov., sp. nov., an anaerobic, amino-acid-degrading bacterium from soil of a Japanese rice field. Int. J. Syst. Evol. Microbiol. 2013, 63, 3679–3686. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Wang, Z.; Wang, Q.; Zhu, C.; Wu, Z. An anaerobic dynamic membrane bioreactor (AnDMBR) for landfill leachate treatment: Performance and microbial community identification. Bioresour. Technol. 2014, 161, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Henze, M.; van Loosdrecht, M.C.; Ekama, G.A.; Brdjanovic, D. Biological Wastewater Treatment: Principles, Modelling and Design; IWA Pub: London, UK, 2008. [Google Scholar]
- Ketheesan, B.; Stuckey, D.C. Effects of hydraulic/organic shock/transient loads in anaerobic wastewater treatment: A review. Crit. Rev. Environ. Sci. Technol. 2015, 45, 2693–2727. [Google Scholar] [CrossRef]
- Ziganshin, A.M.; Liebetrau, J.; Pröter, J.; Kleinsteuber, S. Microbial community structure and dynamics during anaerobic digestion of various agricultural waste materials. Appl. Microbiol. Biotechnol. 2013, 97, 5161–5174. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.-H.; Park, J.-H.; Kang, H.-J.; Lee, Y.H.; Lee, T.J.; Park, H.-D. Distribution and abundance of Spirochaetes in full-scale anaerobic digesters. Bioresour. Technol. 2013, 145, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Conklin, A.; Stensel, H.D.; Ferguson, J. Growth kinetics and competition between Methanosarcina and Methanosaeta in mesophilic anaerobic digestion. Water Environ. Res. 2006, 78, 486–496. [Google Scholar] [CrossRef] [PubMed]
- Ariesyady, H.D.; Ito, T.; Okabe, S. Functional bacterial and archaeal community structures of major trophic groups in a full-scale anaerobic sludge digester. Water Res. 2007, 41, 1554–1568. [Google Scholar] [CrossRef] [PubMed]
- Meabe, E.; Déléris, S.; Soroa, S.; Sancho, L. Performance of anaerobic membrane bioreactor for sewage sludge treatment: Mesophilic and thermophilic processes. J. Membr. Sci. 2013, 446, 26–33. [Google Scholar] [CrossRef]
- Regueiro, L.; Carballa, M.; Lema, J.M. Outlining microbial community dynamics during temperature drop and subsequent recovery period in anaerobic co-digestion systems. Bioresour. Technol. 2014, 192, 179–186. [Google Scholar] [CrossRef] [PubMed]
- De Vrieze, J.; Hennebel, T.; Boon, N.; Verstraete, W. Methanosarcina: The rediscovered methanogen for heavy duty biomethanation. Bioresour. Technol. 2012, 112, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Dar, S.A.; Kleerebezem, R.; Stams, A.J.; Kuenen, J.G.; Muyzer, G. Competition and coexistence of sulfate-reducing bacteria, acetogens and methanogens in a lab-scale anaerobic bioreactor as affected by changing substrate to sulfate ratio. Appl. Microbiol. Biotechnol. 2008, 78, 1045–1055. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- JWH, S.; Elferink, O.; Visser, A.; Pol, H.; Look, W.; Stams, A.J. Sulfate reduction in methanogenic bioreactors. FEMS Microbiol. Rev. 1994, 15, 119–136. [Google Scholar]
- Win, T.T.; Kim, H.; Cho, K.; Song, K.G.; Park, J. Monitoring the microbial community shift throughout the shock changes of hydraulic retention time in an anaerobic moving bed membrane bioreactor. Bioresour. Technol. 2016, 202, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Yin, Q.; Zhu, X.; Zhan, G.; Bo, T.; Yang, Y.; Tao, Y.; He, X.; Li, D.; Yan, Z. Enhanced methane production in an anaerobic digestion and microbial electrolysis cell coupled system with co-cultivation of Geobacter and Methanosarcina. J. Environ. Sci. 2016, 42, 210–214. [Google Scholar] [CrossRef] [PubMed] [Green Version]







| Period | Samples | Sampling Period (d) | COD Feed (mg/L) | COD Effluent (mg/L) | MLSS (g/L) | ORP (mv) | pH | Alkalinity (mg/L CaCO3) |
|---|---|---|---|---|---|---|---|---|
| Startup | A2 | 70 | 455.00 | 173.47 | 6.73 | −160.63 ± 55.75 | 7.72 | 529.00 ± 10.39 |
| B3 | 100 | 475.42 | 100.00 | 5.55 | −174.45 ± 39.07 | 7.90 | ||
| Steady–state period | C4 | 140 | 380.50 | 17.00 | 5.45 | −361.70 ± 30.62 | 7.97 | 675.25 ± 188.42 |
| D5 | 162 | 476 | 14.33 | 4.78 | −310.88 ± 52.98 | 7.96 | ||
| E8 | 190 | 391 | 34.66 | 5.09 | −344.81 ± 26.42 | 7.77 | ||
| Overloading | F10 | 210 | 371.50 | 41.50 | 5.78 | −226.28 ± 23.04 | 7.44 | 537.33 ± 238.28 |
| J15 | 230 | 500.80 | 153.0 | 5.89 | −261.62 ± 37.55 | 7.40 | ||
| Recovery period | K16 | 245 | 504.12 | 76.50 | 6.22 | −369.75 ± 29.90 | 7.29 | 758.00 ± 136.53 |
| L18 | 250 | 470.90 | 52.50 | 6.76 | −364.00 ± 36.13 | 7.12 | ||
| M20 | 260 | 509.58 | 42.66 | 5.33 | −373.41 ± 30.20 | 7.36 | ||
| N23 | 275 | 421.83 | 43.08 | 5.11 | −387.69 ± 15.49 | 7.43 | ||
| P24 | 285 | 582.33 | 43.83 | 4.77 | −373.50 ± 14.00 | 7.26 |
| Phyla and Genera Classification | Seed | Start-Up | Steady-State | Overloading | Recovery Operation | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Phyla | Genera | S1 | S2 | A2 | B3 | C4 | D5 | E8 | F10 | J15 | K16 | L18 | M20 | N23 | P24 |
| Proteobacteria | Desulfovibrio | 0.22 | 0.08 | 0.23 | 0.13 | 0.46 | 0.05 | 0.41 | 0.21 | 0.06 | 0.23 | 1.03 | 0.42 | 0.71 | 1.21 |
| Sulfurovum | 1.73 | 0.00 | 0.13 | 0.24 | 0.73 | 0.12 | 0.69 | 0.17 | 0.09 | 0.10 | 0.29 | 0.25 | 0.31 | 0.32 | |
| Comamonas | 0.00 | 4.58 | 0.79 | 0.29 | 0.44 | 0.05 | 1.01 | 0.87 | 2.53 | 1.88 | 0.93 | 1.24 | 0.65 | 0.87 | |
| Acinetobacter | 0.05 | 0.24 | 0.03 | 0.41 | 0.06 | 0.38 | 0.11 | 2.28 | 0.53 | 0.22 | 0.01 | 0.02 | 0.01 | 0.06 | |
| Geobacter | 1.62 | 0.00 | 1.93 | 1.56 | 0.52 | 0.09 | 0.53 | 0.12 | 0.06 | 0.10 | 1.49 | 1.05 | 0.82 | 1.36 | |
| Pseudomonas | 0.01 | 0.03 | 0.07 | 0.08 | 2.26 | 1.35 | 7.72 | 0.36 | 0.03 | 0.04 | 0.01 | 0.01 | 0.01 | 0.03 | |
| Bacteroidete | Bacteroides | 0.04 | 0.11 | 0.00 | 0.01 | 0.05 | 0.01 | 0.07 | 17.04 | 0.15 | 0.06 | 0.11 | 0.03 | 0.13 | 0.24 |
| Firmicutes | Acidaminococcus | 0.00 | 0.00 | 0.00 | 0.00 | 0.02 | 0.00 | 0.01 | 2.00 | 0.05 | 0.00 | 0.00 | 0.01 | 0.00 | 0.00 |
| Clostridium sensu stricto | 0.56 | 0.12 | 0.03 | 0.02 | 0.03 | 0.00 | 0.01 | 2.08 | 1.74 | 0.89 | 0.61 | 1.41 | 0.56 | 0.53 | |
| Syntrophomonas | 0.00 | 1.30 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.000 | 1.71 | 0.92 | 0.03 | 0.47 | 0.06 | 0.05 | |
| Synergistete | Aminivibrio | 7.22 | 0.00 | 0.87 | 0.39 | 1.16 | 0.06 | 1.57 | 0.32 | 0.15 | 0.11 | 0.05 | 0.12 | 0.04 | 0.05 |
| Aminiphilus | 2.37 | 0.00 | 0.17 | 0.02 | 0.25 | 0.00 | 0.33 | 0.04 | 0.01 | 0.00 | 0.00 | 0.02 | 0.00 | 0.00 | |
| Acidobacteria | Gp17 | 0.00 | 0.00 | 0.03 | 0.04 | 0.37 | 1.87 | 0.26 | 0.04 | 0.01 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| Gp6 | 0.00 | 0.02 | 0.09 | 0.08 | 0.41 | 0.92 | 0.21 | 0.16 | 0.06 | 0.05 | 0.03 | 0.03 | 0.01 | 0.02 | |
| Candidatus Saccharibacteria | Saccharibacteria | 0.03 | 1.28 | 1.62 | 1.07 | 0.49 | 0.08 | 0.39 | 0.22 | 1.02 | 1.15 | 0.70 | 0.85 | 0.86 | 0.70 |
| Euryarchaeota | Methanolin-ea | 4.68 | 0.02 | 1.32 | 0.87 | 5.24 | 1.58 | 6.45 | 0.92 | 0.94 | 1.28 | 0.77 | 0.88 | 0.85 | 0.76 |
| Methanothrix | 6.29 | 0.92 | 0.93 | 0.50 | 3.07 | 1.36 | 2.12 | 0.46 | 2.05 | 1.87 | 2.34 | 2.33 | 2.35 | 1.71 | |
| Methanobac-terium | 5.51 | 7.06 | 0.71 | 0.10 | 0.59 | 0.28 | 0.47 | 0.23 | 1.21 | 1.16 | 0.92 | 0.86 | 0.64 | 0.80 | |
| Methanospi-rillum | 0.31 | 0.29 | 0.03 | 0.02 | 0.37 | 0.06 | 0.44 | 0.04 | 0.72 | 0.76 | 0.24 | 0.38 | 0.28 | 0.28 | |
| Methanomethylovorans | 0.28 | 0.00 | 0.18 | 0.09 | 0.10 | 0.11 | 0.08 | 0.02 | 0.02 | 0.01 | 0.00 | 0.00 | 0.00 | 0.01 | |
| Methanocorpusculum | 0.00 | 0.00 | 0.00 | 0.00 | 0.03 | 0.00 | 0.04 | 0.19 | 0.01 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | |
| Methanobrevibacter | 0.00 | 0.03 | 0.00 | 0.00 | 0.00 | 0.00 | 0.01 | 0.55 | 0.12 | 0.12 | 0.02 | 0.04 | 0.01 | 0.03 | |
| No. | Days | Number of Sequences | OTU | Chao | Ace | Coverage | Shannon | Simpson |
|---|---|---|---|---|---|---|---|---|
| Seed1 | 0 | 29,443 | 609 | 743.10 | 745.92 | 0.994 | 4.199 | 0.034 |
| A2 | 70 | 29,487 | 1108 | 1331.54 | 1341.14 | 0.990 | 5.054 | 0.019 |
| B3 | 100 | 29,660 | 1017 | 1231.55 | 1257.01 | 0.990 | 4.628 | 0.035 |
| C4 | 140 | 29,107 | 1073 | 1388.16 | 1392.34 | 0.988 | 4.790 | 0.025 |
| D5 | 162 | 29,597 | 679 | 866.62 | 874.25 | 0.993 | 3.816 | 0.094 |
| E8 | 190 | 29,535 | 1002 | 1360.16 | 1355.55 | 0.988 | 4.519 | 0.035 |
| F10 | 210 | 29,230 | 1211 | 1536.00 | 1493.78 | 0.988 | 5.060 | 0.032 |
| Seed2 | 220 | 29,244 | 1041 | 1319.10 | 1320.80 | 0.989 | 4.182 | 0.083 |
| J15 | 230 | 29,372 | 1399 | 1802.48 | 1810.62 | 0.985 | 5.275 | 0.021 |
| K16 | 245 | 29,452 | 1305 | 1652.04 | 1673.08 | 0.986 | 5.111 | 0.027 |
| L18 | 250 | 29,622 | 995 | 1287.90 | 1354.73 | 0.989 | 4.953 | 0.015 |
| M20 | 260 | 29,645 | 1088 | 1409.52 | 1438.39 | 0.988 | 4.841 | 0.029 |
| N23 | 275 | 29,532 | 924 | 1205.00 | 1221.05 | 0.990 | 4.728 | 0.023 |
| P24 | 285 | 29,553 | 993 | 1270.44 | 1313.02 | 0.989 | 4.917 | 0.017 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martin Vincent, N.; Wei, Y.; Zhang, J.; Yu, D.; Tong, J. Characterization and Dynamic Shift of Microbial Communities during Start-Up, Overloading and Steady-State in an Anaerobic Membrane Bioreactor. Int. J. Environ. Res. Public Health 2018, 15, 1399. https://doi.org/10.3390/ijerph15071399
Martin Vincent N, Wei Y, Zhang J, Yu D, Tong J. Characterization and Dynamic Shift of Microbial Communities during Start-Up, Overloading and Steady-State in an Anaerobic Membrane Bioreactor. International Journal of Environmental Research and Public Health. 2018; 15(7):1399. https://doi.org/10.3390/ijerph15071399
Chicago/Turabian StyleMartin Vincent, Nsanzumukiza, Yuansong Wei, Junya Zhang, Dawei Yu, and Juan Tong. 2018. "Characterization and Dynamic Shift of Microbial Communities during Start-Up, Overloading and Steady-State in an Anaerobic Membrane Bioreactor" International Journal of Environmental Research and Public Health 15, no. 7: 1399. https://doi.org/10.3390/ijerph15071399
APA StyleMartin Vincent, N., Wei, Y., Zhang, J., Yu, D., & Tong, J. (2018). Characterization and Dynamic Shift of Microbial Communities during Start-Up, Overloading and Steady-State in an Anaerobic Membrane Bioreactor. International Journal of Environmental Research and Public Health, 15(7), 1399. https://doi.org/10.3390/ijerph15071399