Crude Oil Degrading Fingerprint and the Overexpression of Oxidase and Invasive Genes for n-hexadecane and Crude Oil Degradation in the Acinetobacter pittii H9-3 Strain
Abstract
:1. Introduction
2. Materials and Methods
2.1. Enrichment, Isolation, and Identification of Crude Oil-Degrading Bacteria
2.2. Crude Oil Degradation by Strain H9-3
2.3. Utilization of Other Carbon Sources by A. pittii H9-3
2.4. Differential Expression of Alkane-Degrading Genes in Strain H9-3 Cells under Different Carbon Source Culture Conditions
3. Results
3.1. Isolation, Characterization, and Identification of Crude Oil-Degrading Strains
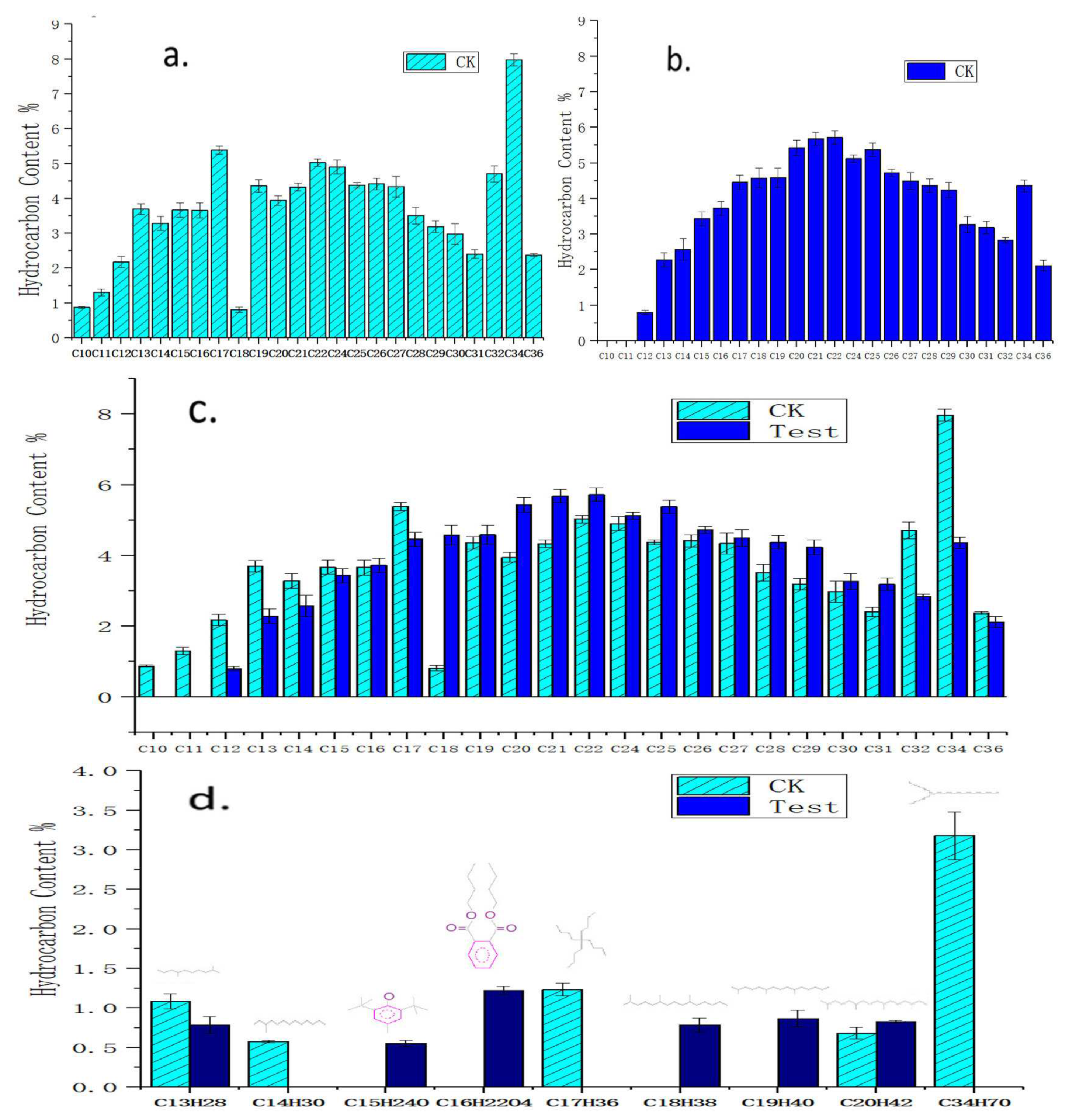
3.2. Degradation Components for Crude Oil by Acinetobacter pittii H9-3
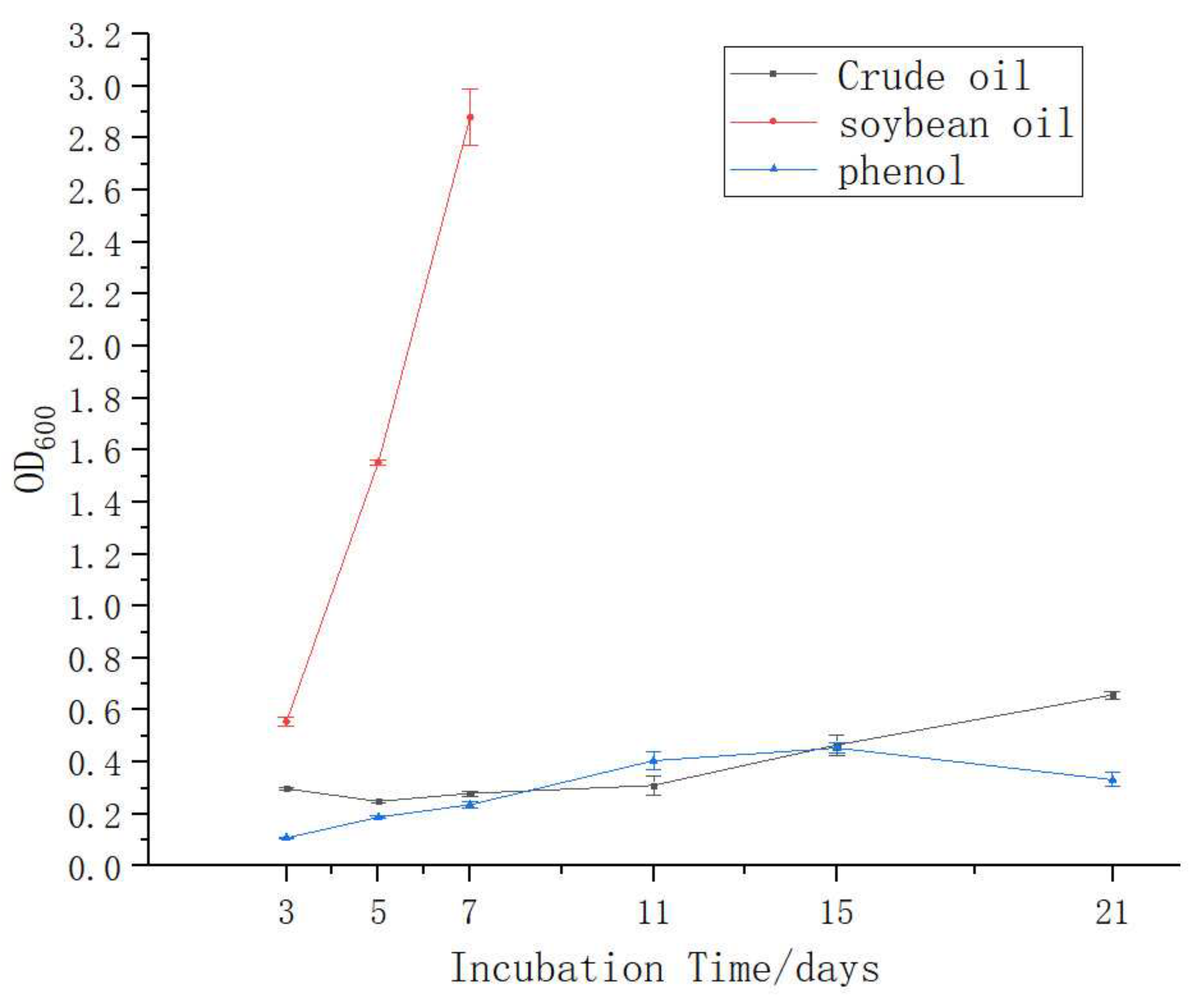
3.3. Growth of A. pittii H9-3 in Different Carbon Sources
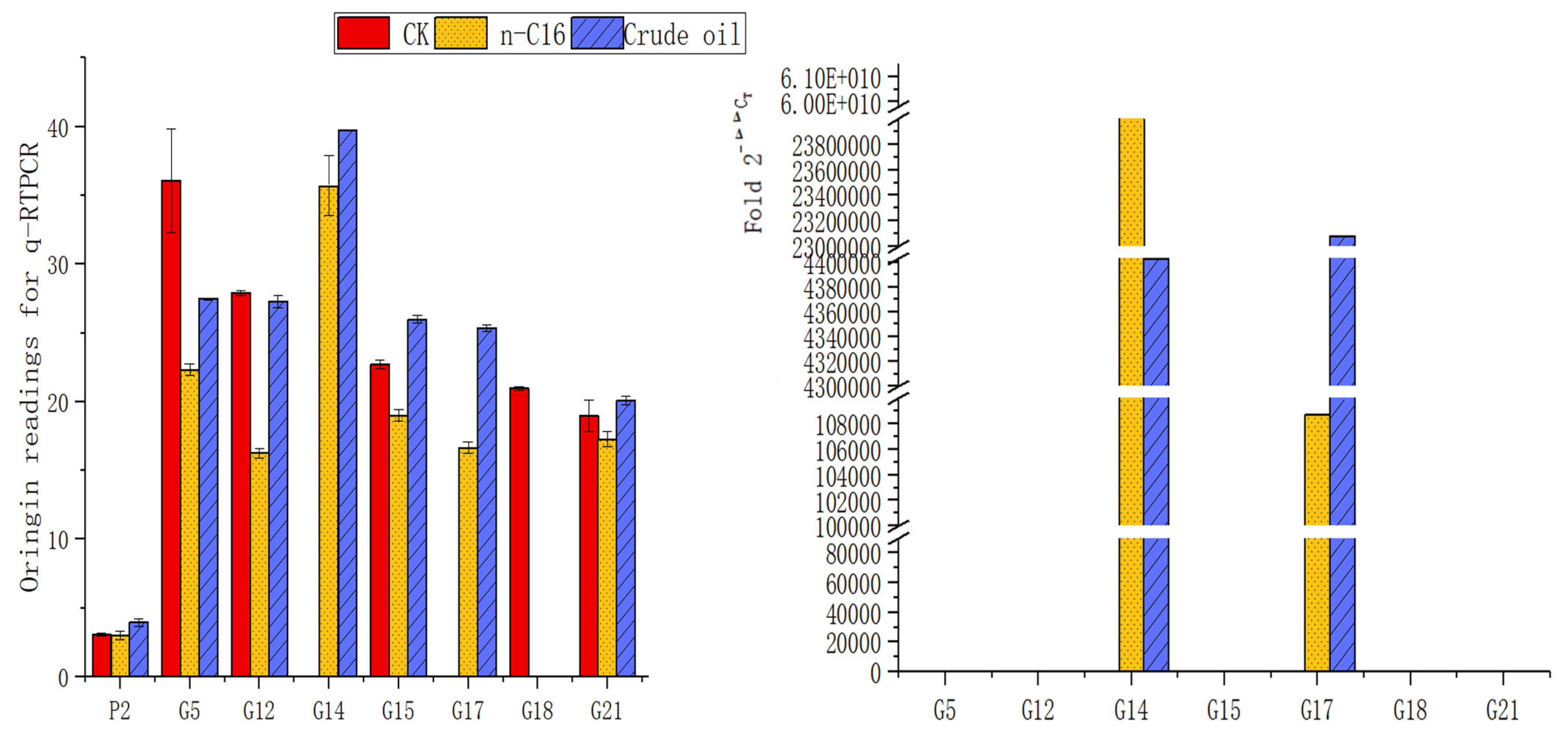
3.4. The Different Expression of Alkane-Degrading Genes in the H9-3 Strain Cultured with Different Carbon Sources
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Head, I.M.; Jones, D.M.; Röling, W.F.M. Marine microorganisms make a meal of oil. Nat. Rev. Microbiol. 2006, 4, 173–182. [Google Scholar] [CrossRef]
- Tay, S.T.; Moy, B.Y.; Maszenan, A.M.; Tay, J.H. Comparing activated sludge and aerobic granules as microbial inocula for phenol biodegradation. Appl. Microbiol. Biotechnol. 2005, 67, 708–713. [Google Scholar] [CrossRef] [PubMed]
- Benyahia, F.; Abdulkarim, M.; Zekri, A.; Chaalal, O.; Hasanain, H. Bioremediation of crude oil contaminated soils: A black art or an engineering challenge? Process. Saf. Environ. Prot. 2005, 83, 364–370. [Google Scholar] [CrossRef]
- Selivanovskaya, S.Y.; Gumerova, R.K.; Galitskaya, P.Y. Assessing the efficiency of methods for the bioremediation of oil production wastes. Contemp. Probl. Ecol. 2013, 6, 542–548. [Google Scholar] [CrossRef]
- Asperger, O.; Kleber, H.P. Metabolism of Alkanes by Acinetobacter; Springer US: New York, NY, USA, 1991; pp. 323–350. [Google Scholar]
- Prince, R.C. Bioremediation of marine oil spills. In Handbook of Hydrocarbon and Lipid Microbiology; Timmis, K.N., Ed.; Springer: Berlin/Heidelberg, Germany, 2010; pp. 2617–2630. [Google Scholar] [CrossRef]
- Muthukamalam, S.; Sivagangavathi, S.; Dhrishya, D.; Sudha, R.S. Characterization of dioxygenases and biosurfactants produced by crude oil degrading soil bacteria. Braz. J. Microbiol. 2017, 48, 637–647. [Google Scholar] [CrossRef] [PubMed]
- Mani, P.; Sivakumar, P.; Balan, S.S. Economic production and oil recovery efficiency of a lipopeptide biosurfactant from a novel marine bacterium Bacillus simplex. Achiev. Life Sci. 2016, 10, 102–110. [Google Scholar] [CrossRef]
- Chandankere, R.; Yao, J.; Cai, M.; Masakorala, K.; Jain, A.K.; Choi, M.M.F. Properties and characterization of biosurfactant in crude oil biodegradation by bacterium Bacillus methylotrophicus USTBa. Fuel 2014, 122, 140–148. [Google Scholar] [CrossRef]
- Romanowska, I.; Strzelecki, B.; Bielecki, S. Biosolubilization of Polish brown coal by Gordonia alkanivorans S7 and Bacillus mycoides NS1020. Fuel Process. Technol. 2015, 131, 430–436. [Google Scholar] [CrossRef]
- Alsaleh, E.; Akbar, A. Occurrence of Pseudomonas aeruginosa in Kuwait soil. Chemosphere 2015, 120, 100–107. [Google Scholar] [CrossRef]
- Pasumarthi, R.; Chandrasekaran, S.; Mutnuri, S. Biodegradation of crude oil by Pseudomonas aeruginosa and Escherichia fergusonii isolated from the Goan coast. Mar. Pollut. Bull. 2013, 76, 276–282. [Google Scholar] [CrossRef]
- Zhou, X.; Xin, Z.J.; Lu, X.H.; Yang, X.P.; Zhao, M.R.; Wang, L.; Liang, J.P. High efficiency degradation crude oil by a novel mutant irradiated from Dietzia strain by 12C6+ heavy ion using response surface methodology. Bioresour. Technol. 2013, 137, 386–393. [Google Scholar] [CrossRef]
- Liu, Y.; Hu, X.; Liu, H. Industrial-scale culturing of the crude oil-degrading marine Acinetobacter sp. strain HC8-3S. Int. Biodeterior. Biodegradation 2016, 107, 56–61. [Google Scholar] [CrossRef]
- Marín, M.; Pedregosa, A.; Ríos, S.; Ortiz, M.L.; Laborda, F. Biodegradation of diesel and heating oil by Acinetobacter calcoaceticus MM5: its possible applications on bioremediation. Int. Biodeterior. Biodegradation 1995, 33, 290–291. [Google Scholar]
- Marín, M.; Pedregosa, A.; Ríos, S.; Laborda, F. Study of factors influencing the degradation of heating oil by Acinetobacter calcoaceticus MM5. Int. Biodeterior. Biodegradation 1996, 38, 69–75. [Google Scholar] [CrossRef]
- Ward, O.P.; Singh, A.; Hamme, J.D.V.; Voordouw, G. Petroleum microbiology. Encyc. Microbiol. 2009, 5, 443–456. [Google Scholar]
- Hassanshahian, M.; Emtiazi, G.; Cappello, S. Isolation and characterization of crude-oil-degrading bacteria from the Persian Gulf and the Caspian Sea. Mar. Pollut. Bull. 2012, 64, 7–12. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Xie, W.; Li, D.; Peng, Y.; Li, Z.; Liu, S. Biodegradation of phenol by bacteria strain Acinetobacter calcoaceticus PA isolated from phenolic wastewater. Int. J. Environ. Res. Public Health 2016, 13, 300. [Google Scholar] [CrossRef]
- Cordova-Rosa, S.M.; Dams, R.I.; Cordova-Rosa, E.V.; Radetski, M.R.; Corrêa, A.X.R.; Radetski, C.M. Remediation of phenol-contaminated soil by a bacterial consortium and Acinetobacter calcoaceticus isolated from an industrial wastewater treatment plant. J. Hazard. Mater. 2009, 164, 61–66. [Google Scholar] [CrossRef]
- Zhan, Y.; Yu, H.; Yan, Y.; Ping, S.; Lu, W.; Zhang, W.; Chen, M.; Lin, M. Benzoate catabolite repression of the phenol degradation in Acinetobacter calcoaceticus PHEA-2. Curr. Microbiol. 2009, 59, 368–373. [Google Scholar] [CrossRef] [PubMed]
- Zhan, Y.; Yan, Y.; Zhang, W.; Yu, H.; Chen, M.; Lu, W.; Ping, S.; Peng, Z.; Yuan, M.; Zhou, Z.; Elmerich, C.; Lin, M. Genome sequence of Acinetobacter calcoaceticus PHEA-2, isolated from industry wastewater. J. Bacteriol. 2011, 193, 2672–2673. [Google Scholar] [CrossRef] [PubMed]
- Nemec, A.; Radolfova-Krizova, L.; Maixnerova, M.; Vrestiakova, E.; Jezek, P.; Sedo, O. Taxonomy of haemolytic and/or proteolytic strains of the genus Acinetobacter with the proposal of Acinetobacter courvalinii sp. nov. (genomic species 14 sensu Bouvet and Jeanjean), Acinetobacter dispersus sp. nov. (genomic species 17), Acinetobacter modestus sp. nov., Acinetobacter proteolyticus sp. nov. and Acinetobacter vivianii sp. nov. Int. J. Syst. Evol. Microbiol. 2016, 66, 1673–1685. [Google Scholar] [PubMed]
- De Vos, D.; Pirnay, J.P.; Bilocq, F.; Jennes, S.; Verbeken, G.; Rose, T. Molecular epidemiology and clinical impact of Acinetobacter calcoaceticus-baumannii complex in a Belgian burn wound center. PLoS ONE 2016, 11, e0156237. [Google Scholar] [CrossRef] [PubMed]
- Feng, M.G.; Pu, X.Y. Time–concentration–mortality modeling of the synergistic interaction of Beauveria bassiana and imidacloprid against Nilaparvata lugens. Pest Manag. Sci. 2005, 61, 363–370. [Google Scholar] [CrossRef] [PubMed]
- Nemec, A.; Krizova, L.; Maixnerova, M.; van der Reijden, T.J.; Deschaght, P.; Passet, V.; Vaneechoutte, M.; Brisse, S.; Dijkshoorn, L. Genotypic and phenotypic characterization of the Acinetobacter calcoaceticus-Acinetobacter baumannii complex with the proposal of Acinetobacter pittii sp. nov. (formerly Acinetobacter genomic species 3) and Acinetobacter nosocomialis sp. nov. (formerly Acinetobacter genomic species 13TU). Res. Microbiol. 2011, 162, 393–404. [Google Scholar] [CrossRef]
- Zhao, L.; Wang, F.; Zhao, J. Identification and functional characteristics of chlorpyrifos-degrading and plant growth promoting bacterium Acinetobacter calcoaceticus. J. Basic Microbiol. 2014, 54, 457–463. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.H.; Ha, S.M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef]
- Wang, X.; Sun, P.; Zhou, Q.; Li, M.; Cao, L.; Zhao, Y. Oil fingerprint analysis of saturated hydrocarbon by internal standard method. Chin. J. Anal. Chem. 2007, 35, 1121–1126. [Google Scholar]
- Iasur-Kruh, L.; Hadar, Y.; Minz, D. Isolation and bioaugmentation of an estradiol-degrading bacterium and its integration into a mature biofilm. Appl. Environ. Microbiol. 2011, 77, 3734–3740. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Sun, J.Q.; Xu, L.; Tang, Y.Q.; Chen, F.M.; Wu, X.L. Simultaneous degradation of phenol and n-hexadecane by Acinetobacter strains. Bioresour. Technol. 2012, 123, 664–668. [Google Scholar] [CrossRef]
- Mousavian, S.S.; Rahimi, K.Y. Emulsan production by Acinetobacter calcoaceticus RAG-1 ATCC-31012. Iran. J. Food. Sci. Technol. 2010, 7, 117–125. [Google Scholar]
- Rocha, M.V.P.; Oliveira, A.H.S.; Souza, M.C.M.; Gonçalves, L.R.B. Natural cashew apple juice as fermentation medium for biosurfactant production by Acinetobacter calcoaceticus. World J. Microbiol. Biotechnol. 2006, 22, 1295–1299. [Google Scholar] [CrossRef]
- Karlsson, A.; Beharry, Z.M.; Matthew, E.D.; Coulter, E.D.; Neidle, E.L.; Jr, K.D.; Eklund, H.; Ramaswamy, S. X-ray crystal structure of benzoate 1,2-dioxygenase reductase from Acinetobacter sp. strain ADP1. J. Mol. Biol. 2002, 318, 261–272. [Google Scholar] [CrossRef]




| Code | Primer Sequence | Length (bp) | Gene ID |
|---|---|---|---|
| P2 | f-CGGCTTTTTGAGATTAGCATC- | 188 | * |
| r-CGCAACCCTTTTCCTTATTTG- | |||
| G5 | f-CAGCCAATAAAGGTCGTAGCA | 112 | gi|325121063|gb|CP002177.1|:3263986-3265761 |
| r-CGGAAGTCAATAGCGTCTGTC | |||
| G12 | f-TGGGACGACGACGATAGATTA- | 212 | gi|325121063|gb|CP002177.1|:c510669-510157 |
| r-TTCCAGTTAAAGCGAACAGTGA- | |||
| G14 | f-AAGAAACTGCTGCTGAACACG- | 233 | gi|325121063|gb|CP002177.1|:210587-211186 |
| r-CCCGTTGGGTTGAATACCTA- | |||
| G15 | f-ACCCTATCTGACGCAGCCTAT- | 216 | gi|325121063|gb|CP002177.1|:c510104-509088 |
| r-TTGAATCTGGAATACCGCATC- | |||
| G17 | f-TGGAGATGAAGTTGAGGCAAT- | 129 | gi|325121063|gb|CP002177.1|:c2325664-2318453 |
| r-GCTGGTGTGCTGTCGTTAGTT- | |||
| G18 | f-CGCTGAAAGCTATCGTGAAAT- | 108 | gi|325121063|gb|CP002177.1|:2835719-2835901 |
| r-GCGATTTCTGCTAATTCTTCG- | |||
| G21 | f-GCCAGCCAAACCCATTATTAC- | 242 | gi|325121063|gb|CP002177.1|:c508226-507033 |
| r-CTGCCACCAACTCTTTAGGAA- |
| Carbon Source | (The Values of Optical Density at 600 nm) | ||
|---|---|---|---|
| 3 days | 5 days | 7 days | |
| Crude oil | 0.30 | 0.25 | 0.28 |
| Soybean oil | 0.56 | 1.55 | 2.88 |
| Phenols | 0.11 | 0.19 | 0.24 |
| -Naphthol | 0.06 | ND | ND |
| Aniline | 0.02 | ND | ND |
| Methylbenzene | 0 | ND | ND |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Wang, Q.; Liu, L. Crude Oil Degrading Fingerprint and the Overexpression of Oxidase and Invasive Genes for n-hexadecane and Crude Oil Degradation in the Acinetobacter pittii H9-3 Strain. Int. J. Environ. Res. Public Health 2019, 16, 188. https://doi.org/10.3390/ijerph16020188
Wang Y, Wang Q, Liu L. Crude Oil Degrading Fingerprint and the Overexpression of Oxidase and Invasive Genes for n-hexadecane and Crude Oil Degradation in the Acinetobacter pittii H9-3 Strain. International Journal of Environmental Research and Public Health. 2019; 16(2):188. https://doi.org/10.3390/ijerph16020188
Chicago/Turabian StyleWang, Yang, Qiuyu Wang, and Limei Liu. 2019. "Crude Oil Degrading Fingerprint and the Overexpression of Oxidase and Invasive Genes for n-hexadecane and Crude Oil Degradation in the Acinetobacter pittii H9-3 Strain" International Journal of Environmental Research and Public Health 16, no. 2: 188. https://doi.org/10.3390/ijerph16020188
APA StyleWang, Y., Wang, Q., & Liu, L. (2019). Crude Oil Degrading Fingerprint and the Overexpression of Oxidase and Invasive Genes for n-hexadecane and Crude Oil Degradation in the Acinetobacter pittii H9-3 Strain. International Journal of Environmental Research and Public Health, 16(2), 188. https://doi.org/10.3390/ijerph16020188




