Hydrogen and Water Bonding between Glycosaminoglycans and Phospholipids in the Synovial Fluid: Molecular Dynamics Study
Abstract
:1. Introduction
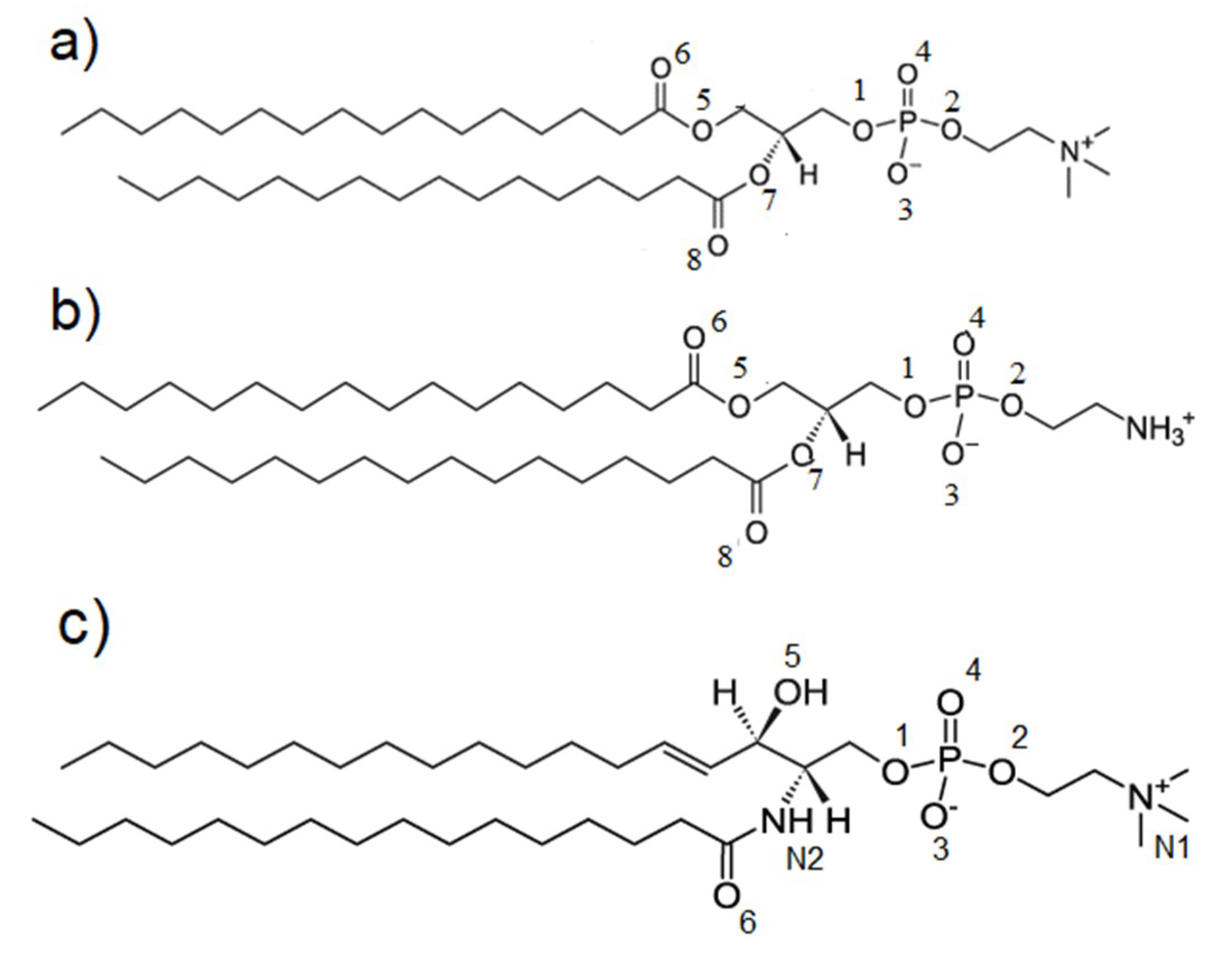
2. Materials and Methods
2.1. Molecular Dynamics Force Field
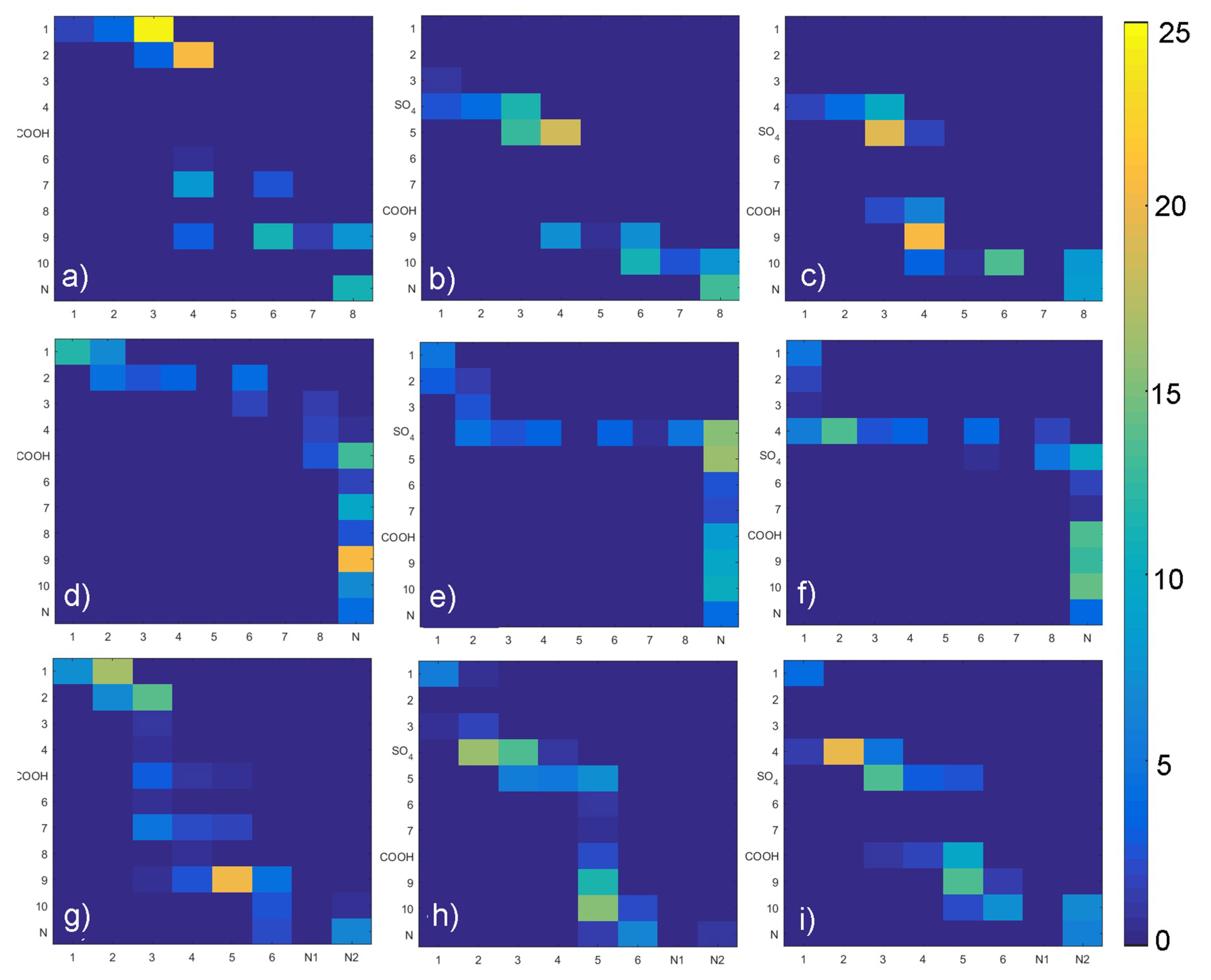
2.2. Hydrogen Bond Identification
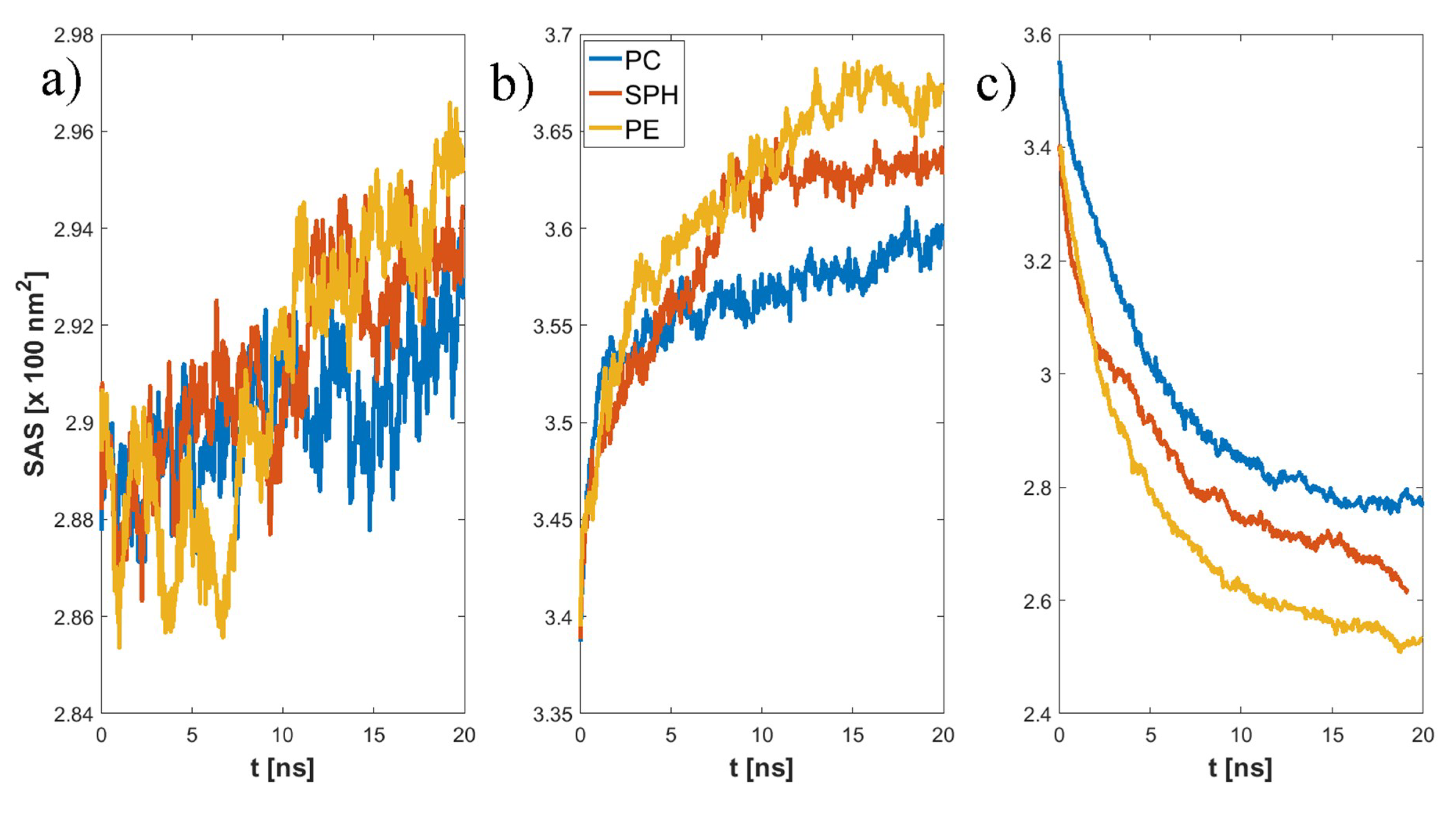
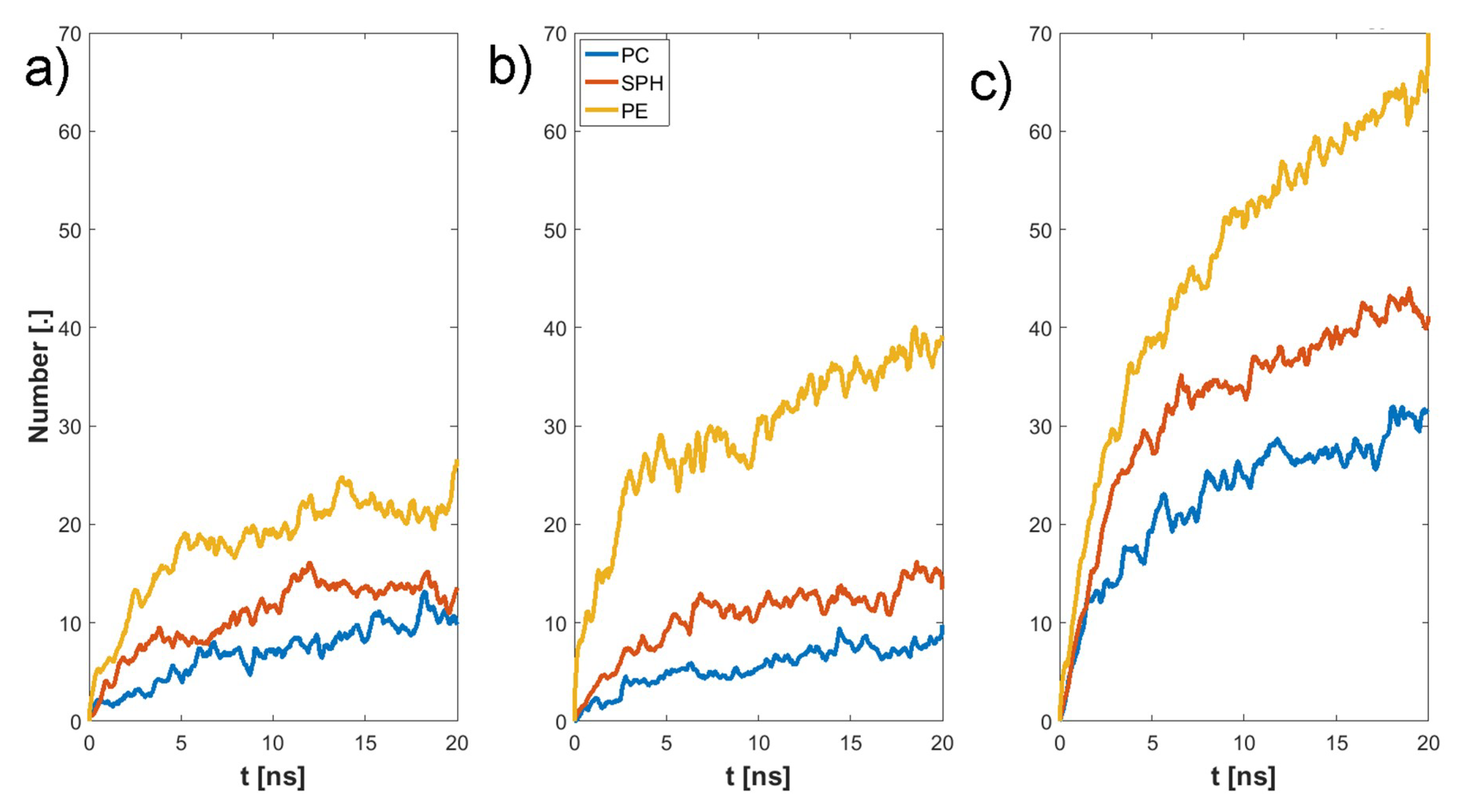
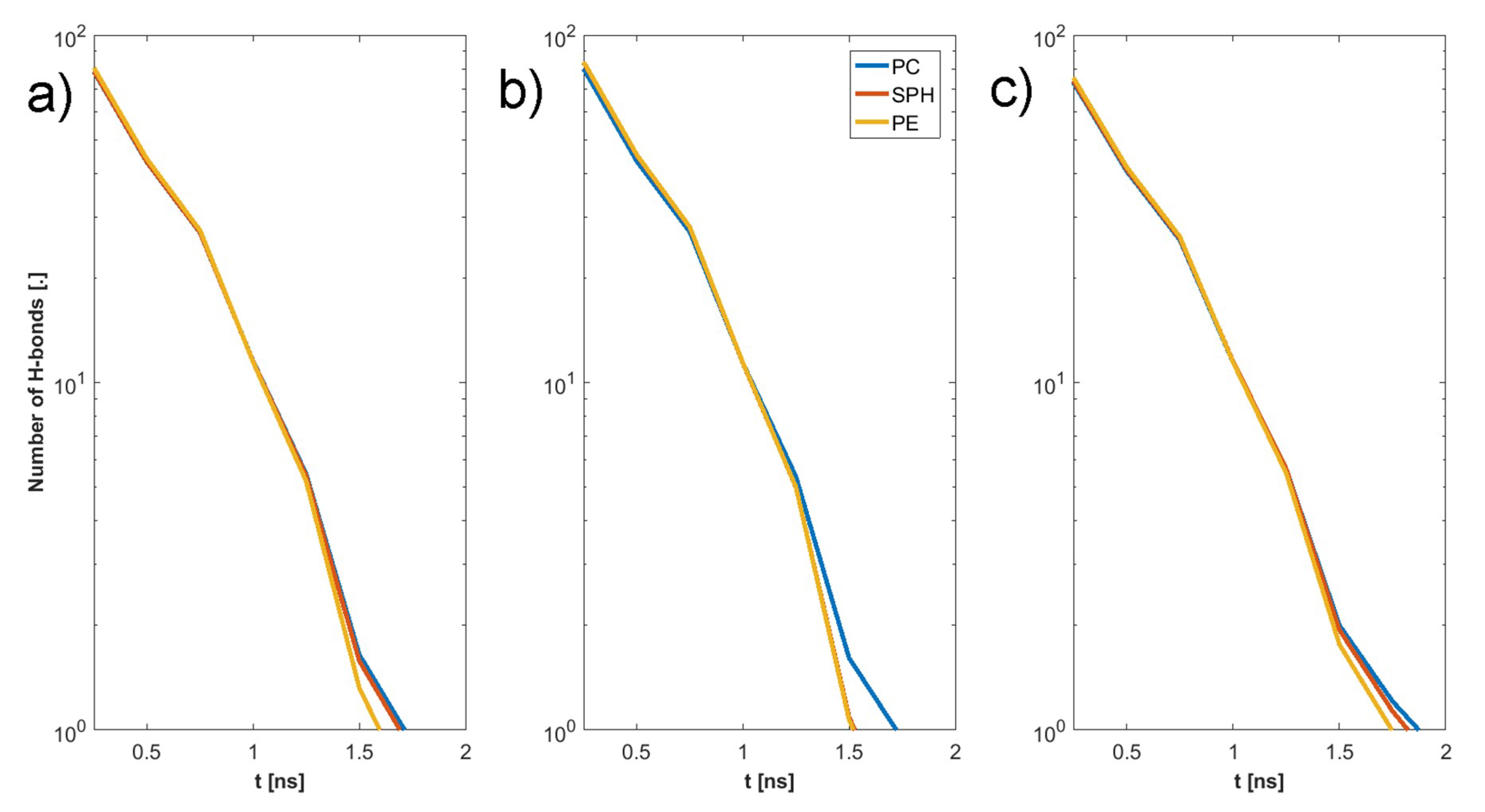
3. Results
4. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Sophia Fox, A.J.; Bedi, A.; Rodeo, S.A. The basic science of articular cartilage: Structure, composition, and function. Sports Health 2009, 1, 461–468. [Google Scholar] [CrossRef]
- Buckwalter, J.A.; Mankin, H.J. Articular cartilage, part 1: Tissue design and chondrocyte-matrix interaction. J. Bone Jt. Surg. Am. 1997, 79, 600–611. [Google Scholar] [CrossRef]
- Mow, V.C.; Ratcliffe, A. Structure and Function of Articular Cartilage and Meniscus, 2nd ed.; Lippincott-Raven: Philadelphia, PA, USA, 1997. [Google Scholar]
- Linn, F.C.; Sokoloff, L. Movement and composition of interstitial fluid of cartilage. Arthritis Rheum. 1965, 8, 481–494. [Google Scholar] [CrossRef]
- Maroudas, A. Physiochemical properties of articular cartilage. In Adult Articular Cartilage; Freeman, M.A.R., Ed.; Pitman Medical Publishing: Kent, UK, 1979; pp. 215–290. [Google Scholar]
- Wierzcholski, K. Joint cartilage lubrication with phospholipid bilayer. Tribologia 2016, 266, 145–157. [Google Scholar] [CrossRef]
- Bełdowski, P.; Weber, P.; Andrysiak, T.; Ii, W.A.; Ledziński, D.; De Leon, T.; Gadomski, A. Anomalous Behavior of Hyaluronan Crosslinking Due to the Presence of Excess Phospholipids in the Articular Cartilage System of Osteoarthritis. Int. J. Mol. Sci. 2017, 18, 2779. [Google Scholar] [CrossRef] [PubMed]
- Buckwalter, J.A.; Martin, J. Degenerative join disease. Clin. Symp. 1995, 47, 1–32. [Google Scholar] [PubMed]
- Cummings, N.A.; Nordby, G.L. Measurement of synovial fluid pH in normal and arthritic knees. Arthritis Rheum. 1966, 9, 47–56. [Google Scholar] [CrossRef]
- Bole, G.G.; Peltier, D.F. Synovial fluid lipids in normal individuals and patients with rheumatoid arthritis. Arthritis Rheum. 1962, 5, 589–601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krieger, E.; Vriend, G. New ways to boost molecular dynamics simulations. J. Comput. Chem. 2015, 36, 996–1007. [Google Scholar] [CrossRef] [PubMed]
- Andrysiak, T.; Bełdowski, P.; Siódmiak, J.; Weber, P.; Ledziński, D. Hyaluronan-Chondroitin Sulphate Anomalous Crosslinking Due to Temperature Changes. Polymers 2018, 10, 560. [Google Scholar] [CrossRef] [PubMed]
- Mark, P.; Nilsson, L. Structure and Dynamics of the TIP3P, SPC, and SPC/E Water Models at 298 K. J. Phys. Chem. A 2001, 105, 9954–9960. [Google Scholar] [CrossRef]
- Berendsen, H.J.C.; Postma, J.P.M.; van Gunsteren, W.F.; DiNola, A.; Haak, J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984, 81, 3684–3690. [Google Scholar] [CrossRef] [Green Version]
- Duan, Y.; Wu, C.; Chowdhury, S.; Lee, M.C.; Xiong, G.; Zhang, W.; Yang, R.; Cieplak, P.; Luo, R.; Lee, T.; et al. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J. Comput. Chem. 2003, 24, 1999–2012. [Google Scholar] [CrossRef] [PubMed]
- Connoly, M.L. Analytical molecular surface calculation. J. Appl. Cryst. 1983, 16, 548–558. [Google Scholar] [CrossRef]
- Richmond, T.J. Solvent accessible surface area and excluded volume in proteins. Analytical equations for overlapping spheres and implications for the hydrophobic effect. J. Mol. Boil. 1984, 178, 63–89. [Google Scholar] [CrossRef]
- Mohammadiarani, H.; Vashisth, H.; Neubig, R.R.; Shaw, V.S. Differential Protein Dynamics of Regulators of G-Protein Signaling: Role in Specificity of Small-Molecule Inhibitors. J. Am. Chem. Soc. 2018, 140, 3454–3460. [Google Scholar]
- Krieger, E.; Dunbrack, R.L., Jr.; Hooft, R.W.; Krieger, B. Assignment of protonation states in proteins and ligands: Combining pKa prediction with hydrogen bonding network optimization. Methods Mol. Biol. 2012, 819, 405–421. [Google Scholar]
- Kwiecinski, J.; Dorosz, S.; Ludwig, T.; Abubacker, S.; Cowman, M.; Schmidt, T. The effect of molecular weight on hyaluronan’s cartilage boundary lubricating ability—Alone and in combination with proteoglycan 4. Osteoarthr. Cartil. 2011, 19, 1356–1362. [Google Scholar] [CrossRef]
- Seror, J.; Zhu, L.; Goldberg, R.; Day, A.J.; Klein, J. Supramolecular synergy in the boundary lubrication of synovial joints. Nat. Commun. 2015, 6, 6497. [Google Scholar] [CrossRef]
- Das, S.; Banquy, X.; Zappone, B.; Greene, G.W.; Jay, G.D.; Israelachvili, J.N. Synergistic Interactions between Grafted Hyaluronic Acid and Lubricin Provide Enhanced Wear Protection and Lubrication. Biomacromolecules 2013, 14, 1669–1677. [Google Scholar] [CrossRef]
- Wieland, D.C.F.; Degen, P.; Gayer, S.; Raj, A.; An, J.; Claesson, P.; Willumeit-Römer, R.; Zander, T.; Dėdinaitė, A. Structure of DPPC–hyaluronan interfacial layers – effects of molecular weight and ion composition. Soft Matter 2016, 12, 729–740. [Google Scholar] [CrossRef] [PubMed]
- Bełdowski, P.; Weber, P.; Dėdinaitė, A.; Claesson, P.M.; Gadomski, A. Correction: Physical crosslinking of hyaluronic acid in the presence of phospholipids in an aqueous nano-environment. Soft Matter 2018, 14, 9730. [Google Scholar] [CrossRef] [PubMed]
- Hari, G.G.; Hales, C.A. Chemistry and Biology of Hyaluronan; Elsevier Science: Amsterdam, The Netherlands, 2008; ISBN 9780080472225. [Google Scholar]
- Kosinska, M.K.; Ludwig, T.E.; Liebisch, G.; Zhang, R.; Siebert, H.-C.; Wilhelm, J.; Kaesser, U.; Dettmeyer, R.B.; Klein, H.; Ishaque, B.; et al. Articular Joint Lubricants during Osteoarthritis and Rheumatoid Arthritis Display Altered Levels and Molecular Species. PLoS ONE 2015, 10. [Google Scholar] [CrossRef] [PubMed]
- Snelling, S.; Rout, R.; Davidson, R.; Clark, I.; Carr, A.; Hulley, P.; Price, A.; Hulley, P. A gene expression study of normal and damaged cartilage in anteromedial gonarthrosis, a phenotype of osteoarthritis. Osteoarthr. Cartil. 2014, 22, 334–343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nishimura, M.; Yan, W.; Mukudai, Y.; Nakamura, S.; Nakamasu, K.; Kawata, M.; Kawamoto, T.; Noshiro, M.; Hamada, T.; Kato, Y. Role of chondroitin sulphate-hyaluronan interactions in the viscoelastic properties of extracellular matrices and fluids. Biochim. Biophys. Acta 1998, 1380, 1–9. [Google Scholar] [CrossRef]
- Band, P.A.; Heeter, J.; Wiśniewski, H.G.; Liublinska, V.; Pattanayak, C.W.; Karia, R.J.; Stabler, T.; Balazs, E.A.; Kraus, V.B. Hyaluronan molecular weight distribution is associated with the risk of knee osteoarthritis progression. Osteoarthr. Cartil. 2015, 23, 70–76. [Google Scholar] [CrossRef]
- Temple-Wong, M.M.; Ren, S.; Quach, P.; Hansen, B.C.; Chen, A.C.; Hasegawa, A.; D’Lima, D.D.; Koziol, J.; Masuda, K.; Lotz, M.K.; et al. Hyaluronan concentration and size distribution in human knee synovial fluid: Variations with age and cartilage degeneration. Arthritis Res. Ther. 2016, 18, 18. [Google Scholar] [CrossRef]
- Lee, D.W.; Banquy, X.; Das, S.; Cadirov, N.; Jay, G.; Israelachvili, J. Effects of molecular weight of grafted hyaluronic acid on wear initiation. Acta Biomater. 2014, 10, 1817–1823. [Google Scholar] [CrossRef]







© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bełdowski, P.; Mazurkiewicz, A.; Topoliński, T.; Małek, T. Hydrogen and Water Bonding between Glycosaminoglycans and Phospholipids in the Synovial Fluid: Molecular Dynamics Study. Materials 2019, 12, 2060. https://doi.org/10.3390/ma12132060
Bełdowski P, Mazurkiewicz A, Topoliński T, Małek T. Hydrogen and Water Bonding between Glycosaminoglycans and Phospholipids in the Synovial Fluid: Molecular Dynamics Study. Materials. 2019; 12(13):2060. https://doi.org/10.3390/ma12132060
Chicago/Turabian StyleBełdowski, Piotr, Adam Mazurkiewicz, Tomasz Topoliński, and Tomasz Małek. 2019. "Hydrogen and Water Bonding between Glycosaminoglycans and Phospholipids in the Synovial Fluid: Molecular Dynamics Study" Materials 12, no. 13: 2060. https://doi.org/10.3390/ma12132060





