Multi Dynamic Extraction: An Innovative Method to Obtain a Standardized Chemically and Biologically Reproducible Polyphenol Extract from Poplar-Type Propolis to Be Used for Its Anti-Infective Properties
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Hydroalcoholic Propolis Extract Preparation
2.3. M.E.D. Propolis Preparation
2.4. Analyses of Hydroalcoholic Propolis Extracts and M.E.D. Propolis by RP-HPLC–PDA–ESI–MSn
2.5. Antimicrobial Assays
2.6. Statistical Analysis
3. Results
3.1. RP-HPLC–PDA–ESI–MSn Analyses of Hydroalcoholic Propolis Extracts
3.2. RP-HPLC–PDA–ESI–MSn Analyses of Non-Ethanolic M.E.D. Propolis
3.3. Antimicrobial Activity of M.E.D. Propolis Extracts
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Pasupuleti, V.R.; Sammugam, L.; Ramesh, N.; Gan, S.H. Honey, propolis, and royal jelly: A comprehensive review of their biological actions and health benefits. Oxid. Med. Cell. Longev. 2017, 2017, 1259510. [Google Scholar] [CrossRef] [PubMed]
- Ghisalberti, E.L. Propolis: A review. Bee World 1979, 60, 59–84. [Google Scholar] [CrossRef]
- Bankova, V.; Bertelli, D.; Borba, R.; Conti, B.J.; da Silva Cunha, I.B.; Danert, C.; Eberlin, M.N.; Falcão, S.I.; Isla, M.I.; Papotti, G.; et al. Standard methods for Apis mellifera propolis research. J. Apic. Res. 2016, 58, 1–49. [Google Scholar] [CrossRef]
- Gómez-Caravaca, A.M.; Gómez-Romero, M.; Arráez-Román, D.; Segura-Carretero, A.; Fernández-Gutiérrez, A. Advances in the analysis of phenolic compounds in products derived from bees. J. Pharm. Biomed. Anal. 2006, 41, 1220–1234. [Google Scholar] [CrossRef]
- Zabaiou, N.; Fouache, A.; Trousson, A.; Baron, S.; Zellagui, A.; Lahouel, M.; Lobaccaro, J.A. Biological properties of propolis extracts: Something new from an ancient product. Chem. Phys. Lipids 2017, 207, 214–222. [Google Scholar] [CrossRef]
- Bankova, V.; Popova, M.; Trusheva, B. Propolis volatile compounds: Chemical diversity and biological activity: A review. Chem. Cent. J. 2014, 8, 28. [Google Scholar] [CrossRef]
- Woisky, R.G.; Salatino, A. Analysis of propolis: Some parameters and procedures for chemical quality control. J. Apic. Res. 1998, 37, 99–105. [Google Scholar] [CrossRef]
- Pietta, P.G.; Gardana, C.; Pietta, A.M. Analytical methods for quality control of propolis. Fitoterapia 2002, 73, S7–S20. [Google Scholar] [CrossRef]
- Kubiliene, L.; Laugaliene, V.; Pavilonis, A.; Maruska, A.; Majiene, D.; Barcauskaite, K.; Kubilius, R.; Kasparaviciene, G.; Savickas, A. Alternative preparation of propolis extracts: Comparison of their composition and biological activities. BMC Complement. Altern. Med. 2015, 15, 156. [Google Scholar] [CrossRef]
- Mello, B.C.B.S.; Petrus, J.C.C.; Hubinger, M.D. Concentration of flavonoids and phenolic compounds in aqueous and ethanolic propolis extracts through nanofiltration. J. Food Eng. 2010, 96, 533–539. [Google Scholar] [CrossRef]
- Zaccaria, V.; Curti, V.; Di Lorenzo, A.; Baldi, A.; Maccario, C.; Sommatis, S.; Mocchi, R.; Daglia, M. Effect of green and brown propolis extracts on the expression levels of microRNAs, mRNAs and proteins, related to oxidative stress and inflammation. Nutrients 2017, 9, 1090. [Google Scholar] [CrossRef] [PubMed]
- Galeotti, F.; Maccari, F.; Fachini, A.; Volpi, N. Chemical composition and antioxidant activity of propolis prepared in different forms and in different solvents useful for finished products. Foods 2018, 7, 41. [Google Scholar] [CrossRef]
- Volpi, N.; Fachini, A. Procedimento Per L’ottenimento di Estratti Integrali di Propoli Ricchi in Polifenoli e Dot, ati di Attività Antibatterica e Sua Applicazione Nella Prevenzione e Trattamento di Processi Infettivi di Origine Batterica. UfficioItalianoBrevetti e Marchi No. 0001425516, 2 February 2017. [Google Scholar]
- Castaldo, S.; Capasso, F. Propolis, an old remedy used in modern Medicine. Fitoterapia 2002, 73, S1–S6. [Google Scholar] [CrossRef]
- Kocot, J.; Kiełczykowska, M.; Luchowska-Kocot, D.; Kurzepa, J.; Musik, I. Antioxidant potential of propolis, bee pollen, and royal jelly: Possible medical application. Oxid. Med. Cell. Longev. 2018, 2018, 7074209. [Google Scholar] [CrossRef] [PubMed]
- Havsteen, B.H. The biochemistry and medical significance of the flavonoids. Pharmacol. Ther. 2002, 96, 67–202. [Google Scholar] [CrossRef]
- Bankova, V. Recent trends and important developments in propolis research. Evid. Based. Complement. Alternat. Med. 2005, 2, 29–32. [Google Scholar] [CrossRef]
- Chee, H.Y. In vitro evaluation of the antifungal activity of propolis extract on Cryptococcus neoformans and Candida albicans. Mycobiology 2002, 30, 93–95. [Google Scholar] [CrossRef]
- Ota, C.; Unterkircher, C.; Fantinato, V.; Shimizu, M.T. Antifungal activity of propolis on different species of Candida. Mycoses 2001, 44, 375–378. [Google Scholar] [CrossRef]
- Kujumgiev, A.; Tsvetkova, I.; Serkedjieva, Y.; Bankova, V.; Christov, R.; Popov, S. Antibacterial, antifungal and antiviral activity of propolis of different geographic origin. J. Ethnopharmacol. 1999, 64, 235–240. [Google Scholar] [CrossRef]
- Miorin, P.L.; Levy Junior, N.C.; Custodio, A.R.; Bretz, W.A.; Marcucci, M.C. Antibacterial activity of honey and propolis from Apis mellifera and Tetragonisca angustula against Staphylococcus aureus. J. Appl. Microbiol. 2003, 95, 913–920. [Google Scholar] [CrossRef]
- EFSA Panel on Dietetic Products, Nutrition and Allergies (NDA). Scientific opinion on the substantiation of health claims related to propolis (ID 1242, 1245, 1246, 1247, 1248, 3184) and flavonoids in propolis (ID 1244, 1644, 1645, 3526, 3527, 3798, 3799) pursuant to Article 13(1) of Regulation (EC) No 1924/2006. EFSA J. 2010, 8, 1810–1826. [Google Scholar] [CrossRef]
- Muli, E.M.; Maingi, J.M. Antibacterial activity of Apis mellifera L. propolis collected in three regions of Kenya. J. Venom. Anim. Toxins Trop. Dis. 2007, 13, 655–663. [Google Scholar] [CrossRef]
- Silva, J.; Rodrigues, S.; Feás, X.; Estevinho, L. Antimicrobial activity, phenolic profile and role in the inflammation of propolis. Food Chem. Toxicol. 2012, 50, 1790–1795. [Google Scholar] [CrossRef] [PubMed]
- Pamplona-Zomenhan, L.C.; Pamplona, B.C.; da Silva, C.B.; Marcucci, M.C.; Mimica, L.M. Evaluation of the in vitro antimicrobial activity of an ethanol extract of Brazilian classified propolis on strains of Staphylococcus aureus. Braz. J. Microbiol. 2011, 42, 1259–1264. [Google Scholar] [CrossRef]
- Fernandes, J.R.A.; Sugizaki, M.F.; Fogo, M.L.; Funari, S.R.C.; Lopes, C.A.M. In vitro activity of propolis against bacterial and yeast pathogens isolated from human infections. J. Venom. Anim. Toxins. 1995, 1, 63–69. [Google Scholar] [CrossRef]
- Pepeljnjak, S.; Kosalec, I. Galangin expresses bactericidal activity against multiple-resistant bacteria: MRSA, Enterococcus spp. and Pseudomonas aeruginosa. FEMS Microbiol. Lett. 2004, 240, 111–116. [Google Scholar] [CrossRef]
- Salomão, K.; Dantas, A.P.; Borba, C.M.; Campos, L.C.; Machado, D.G.; Neto, F.R.A.; de Castro, S.L. Chemical composition and microbicidal activity of extracts from Brazilian and Bulgarian propolis. Lett. Appl. Microbiol. 2004, 38, 87–92. [Google Scholar] [CrossRef]
- Mirzoeva, O.K.; Grishanin, R.N.; Calder, P.C. Antimicrobial action of propolis and some of its components: The effects on growth, membrane potential and motility of bacteria. Microbiol. Res. 1997, 152, 239–246. [Google Scholar] [CrossRef]
- Duarte, S.; Rosalen, P.L.; Hayacibar, M.F.; Cury, J.A.; Bowen, W.H.; Marquis, R.E.; Rehder, V.L.; Sartoratto, A.; Ikegaki, M.; Koo, H. The influence of a novel propolis on mutans streptococci biofilms and caries development in rats. Arch. Oral Biol. 2006, 51, 15–22. [Google Scholar] [CrossRef]
- Savka, M.A.; Dailey, L.; Popova, M.; Mihaylova, R.; Merritt, B.; Masek, M.; Le, P.; Nor, S.R.; Ahmad, M.; Hudson, A.O.; et al. Chemical composition and disruption of quorum sensing signaling in geographically diverse united states propolis. Evid. Based Complement. Altern. Med. 2015, 2015, 472593. [Google Scholar] [CrossRef]
- Cushnie, T.P.T.; Hamilton, V.E.S.; Chapman, D.G.; Taylor, P.W.; Lamb, A.J. Aggregation of Staphylococcus aureus following treatment with the antibacterial flavonol galangin. J. Appl. Microbiol. 2007, 103, 1562–1567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ouyang, J.; Sun, F.; Feng, W.; Xie, Y.; Ren, L.; Chen, Y. Antimicrobial activity of galangin and its effects on murein hydrolases of vancomycin-intermediate Staphylococcus aureus (VISA) strain Mu50. Chemotherapy 2018, 63, 20–28. [Google Scholar] [CrossRef] [PubMed]
- Cushnie, T.P.; Hamilton, V.E.; Lamb, A.J. Assessment of the antibacterial activity of selected flavonoids and consideration of discrepancies between previous reports. Microbiol. Res. 2003, 158, 281–289. [Google Scholar] [CrossRef] [PubMed]
- Jaisinghani, R.N. Antibacterial properties of quercetin. Microbiol. Res. 2017, 8, 6877. [Google Scholar] [CrossRef] [Green Version]
- Soromou, L.W.; Zhang, Y.; Cui, Y.; Wei, M.; Chen, N.; Yang, X.; Huo, M.; Baldé, A.; Guan, S.; Deng, X.; et al. Subinhibitory concentrations of pinocembrin exert anti-Staphylococcus aureus activity by reducingα-toxin expression. J. Appl. Microbiol. 2013, 15, 41–49. [Google Scholar] [CrossRef] [Green Version]
- Biva, I.J.; Ndi, C.P.; Griesser, H.J.; Semple, S.J. Antibacterial constituents of Eremophila alternifolia: An Australian aboriginal traditional medicinal plant. J. Ethnopharmacol. 2016, 182, 1–9. [Google Scholar] [CrossRef]
- Nina, N.; Quispe, C.; Jiménez-Aspee, F.; Theoduloz, C. Antibacterial activity, antioxidant effect and chemical composition of propolis from the Región del Maule, Central Chile. Molecules 2015, 20, 18144–18167. [Google Scholar] [CrossRef] [Green Version]
- Krol, W.; Scheller, S.; Shani, J.; Pietsz, G.; Czuba, Z. Synergistic effect of ethanolic extract of propolis and antibiotics on the growth of Staphylococcus aureus. Arzneimittelforsch 1993, 43, 607–609. [Google Scholar]
- Santos, A.; Bastos, M.; Uzeda, M.; Carvalho, A. Antibacterial activity of Brazilian propolis and fractions against oral anaerobic bacteria. J. Ethnopharmacol. 2002, 80, 1–7. [Google Scholar] [CrossRef]
- Liu, B.; Pop, M. ARDB-antibiotic resistance genes database. Nucleic Acids Res. 2009, 37, D443–D447. [Google Scholar] [CrossRef] [Green Version]
- Campoccia, D.; Montanaro, L.; Speziale, P.; Arciola, C.R. Antibiotic-loaded biomaterials and the risks for the spread of antibiotic resistance following their prophylactic and therapeutic clinical use. Biomaterials 2010, 31, 6363–6377. [Google Scholar] [CrossRef] [PubMed]
- Arciola, C.R.; Radin, L.; Alvergna, P.; Cenni, E.; Pizzoferrato, A. Heparin surface treatment of poly(methylmethacrylate) alters adhesion of a Staphylococcus aureus strain: Utility of bacterial fatty acid analysis. Biomaterials 1993, 14, 1161–1164. [Google Scholar] [CrossRef]
- Tiller, J.C.; Liao, C.J.; Lewis, K.; Klibanov, A.M. Designing surfaces that kill bacteria on contact. Proc. Natl. Acad. Sci. USA 2001, 98, 5981–5985. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bazaka, K.; Jacob, M.V.; Crawford, R.J.; Ivanova, E.P. Efficient surface modification of biomaterial to prevent biofilm formation and the attachment of microorganisms. Appl. Microbiol. Biotechnol. 2012, 95, 299–311. [Google Scholar] [CrossRef] [PubMed]
- Marchese, A.; Arciola, C.R.; Coppo, E.; Barbieri, R.; Barreca, D.; Chebaibi, S.; Sobarzo-Sánchez, E.; Nabavi, S.F.; Nabavi, S.M.; Daglia, M. The natural plant compound carvacrol as an antimicrobial and anti-biofilm agent: Mechanisms, synergies and bio-inspired anti-infective materials. Biofouling 2018, 34, 630–656. [Google Scholar] [CrossRef]
- Russo, N.; Cassinelli, C.; Torre, E.; Morra, M.; Iviglia, G. Improvement of the physical properties of guided bone regeneration membrane from porcine pericardium by polyphenols-rich pomace extract. Materials 2019, 12, 2564. [Google Scholar] [CrossRef] [Green Version]
- Williams, D.F. Challenges with the development of biomaterials for sustainable tissue engineering. Front. Bioeng. Biotechnol. 2019, 7, 127. [Google Scholar] [CrossRef] [Green Version]
- Eskandarinia, A.; Kefayat, A.; Rafienia, M.; Agheb, M.; Navid, S.; Ebrahimpour, K. Cornstarch-based wound dressing incorporated with hyaluronic acid and propolis: In vitro and in vivo studies. Carbohydr. Polym. 2019, 216, 25–35. [Google Scholar] [CrossRef]
- Voss, G.T.; Gularte, M.S.; Vogt, A.G.; Giongo, J.L.; Vaucher, R.A.; Echenique, J.V.Z.; Soares, M.P.; Luchese, C.; Wilhelm, E.A.; Fajardo, A.R. Polysaccharide-based film loaded with vitamin C and propolis: A promising device to accelerate diabetic wound healing. Int. J. Pharm. 2018, 552, 340–351. [Google Scholar] [CrossRef]
- Oliveira, R.N.; McGuinness, G.B.; Rouze, R.; Quilty, B.; Cahill, P.; Soares, G.D.A.; Thiré, R.M.S.M. PVA hydrogels loaded with a Brazilian propolis for burn wound healing applications. J. Appl. Polym. Sci. 2015, 132, 42129. [Google Scholar] [CrossRef]
- Boni, B.O.O.; Lamboni, L.; Souho, T.; Gauthier, M.; Yang, G. Immunomodulation and cellular response to biomaterials: The overriding role of neutrophils in healing. Mater. Horiz. 2019, 6, 1122–1137. [Google Scholar] [CrossRef]
- Kmiotek, M.; Bielinski, D.; Piotrowska, M. Propolis as an antidegradant and biocidal agent for natural rubber. J. Appl. Polym. Sci. 2018, 135, 45911. [Google Scholar] [CrossRef]
- Ong, T.H.; Chitra, E.; Ramamurthy, S.; Ling, C.C.S.; Ambu, S.P.; Davamani, F. Cationic chitosan-propolis nanoparticles alter the zeta potential of S. epidermidis, inhibit biofilm formation by modulating gene expression and exhibit synergism with antibiotics. PLoS ONE 2019, 14, e0213079. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.I.; Pant, H.R.; Sim, H.J.; Lee, K.M.; Kim, C.S. Electrospun propolis/polyurethane composite nanofibers for biomedical applications. Mater. Sci. Eng. C Mater. Biol. Appl. 2014, 44, 52–57. [Google Scholar] [CrossRef] [PubMed]
- Patra, J.K.; Das, G.; Fraceto, L.F.; Campos, E.V.R.; Rodriguez-Torres, M.D.P.; Acosta-Torres, L.S.; Diaz-Torres, L.A.; Grillo, R.; Swamy, M.K.; Sharma, S.; et al. Nano based drug delivery systems: Recent developments and future prospects. J. Nanobiotechnol 2018, 16, 71. [Google Scholar] [CrossRef] [Green Version]
- Reinholz, J.; Landfester, K.; Mailänder, V. The challenges of oral drug delivery via nanocarriers. Drug Deliv. 2018, 25, 1694–1705. [Google Scholar] [CrossRef]
- Arafa, M.G.; Ghalwash, D.; El-Kersh, D.M.; Elmazar, M.M. Propolis-based niosomes as oromuco-adhesive films: A randomized clinical trial of a therapeutic drug delivery platform for the treatment of oral recurrent aphthous ulcers. Sci. Rep. 2018, 8, 18056. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.N.; Weng, M.S.; Wu, C.L.; Lin, J.K. Comparison of radical scavenging activity, cytotoxic effects and apoptosis induction in human melanoma cells by Taiwanese propolis from different sources. Evid. Based Complement. Altern. Med. 2004, 1, 175–185. [Google Scholar] [CrossRef] [Green Version]
- Sadeghi-Aliabadi, H.; Hamzeh, J.; Mirian, M. Investigation of Astragalus honey and propolis extract’s cytotoxic effect on two human cancer cell lines and their oncogen and proapoptotic gene expression profiles. Adv. Biomed. Res. 2015, 4, 42. [Google Scholar] [CrossRef]


| Time (min) | % Eluent A | % Eluent B |
|---|---|---|
| 0 | 85 | 15 |
| 30 | 60 | 40 |
| 65 | 45 | 55 |
| 70 | 38 | 62 |
| 85 | 0 | 100 |
| 90 | 0 | 100 |
| 100 | 85 | 15 |
| 110 | 85 | 15 |
| Microbial Strain | Media | Conditions |
|---|---|---|
| S. aureus methicillin-sensitive ATCC25923 (MSSA)(L1280) S. aureus methicillin-resistant (MRSA) (L4064) S. aureus MSSA + glycopeptide-interMediate resistant (GISA) (L3797) S. aureus MRSA + GISA (L3798) S. aureus clinda-inducible erm(A)+ (ND053410) S. aureus community acquired USA300 MRSA (ND054910) S. aureus MRSA + macrolides-resistant (ND060411) S. hominis ATCC27844 (L323) S. epidermidis (L147; ND052110; ND051710) S. epidermidis teicoplanin-resistant (ND042409) S. capitis MRSA (ND021008) S. xylosus MRSA (ND026108) S. simulans (ND029808) S. haemolyticus MRSA (L1730, ND040809; L1729) E. coli hyperpermeable (G1640) E. coli ATCC25922 (L1281) E. coli hyperpermeable (L4242; L47) P. aeruginosa ATCC27853 (L1367) M. catarrhalis (L3292) A. baumannii (L3030) L. monocytogenes ATCC13932 (L1450) B. cereus ATCC10702 (L85) | Mueller Hinton Agar | Aerobic, 24 h, 37 °C |
| S. pneumoniae penicillin-susceptible (L44) S. pneumoniae penicillin-resistant (L3917) S. pneumoniae clindamycin and erythromycin resistant (L1542) S. pneumoniae macrolide and erythromycin resistant (L1402) | Todd Hewitt Agar | Aerobic, 24 h, 37 °C |
| C. parapsilosis ATCC90018 (L3022) C. parapsilosis ATCC22019 (L4119) C. albicans ATCC24443 (L4120) C. albicans ATCC90028 (L3023) C. guillermondii ATCC6260 (L2065) C. kruzei (L2880) A. niger ATCC10535 (L53) | Sabouraud Dextrose Agar | Aerobic, 48 h, 37 °C |
| G. vaginalis (L1629; L1622; L1630) A. vaginae (ND736; ND737) B. fragilis ATCC25285 (L1011) L. paracasei (L1693) L. plantarum (L19) L. gasseri (ND787) L. acidophilus (ND786) C. difficile (L1365; L1366) C. difficile ATCC17858 (L4013) | Brucella Agar with 5% laked horse blood and 1% hemin and vitamin K | Anaerobic, 72 h, 37 °C |
| N. gonorrhoeae (L1600; L1601; L1599) | Brucella Agar with 5% laked horse blood, 1% hemin and vitamin K and 1% isovitalex | Anaerobic, 72 h, 37 °C |
| C. perfrigens (L4053) C. perfrigens ATCC13124 (L3697) P. acnes ATCC25746 (L1016) | Brucella Agar with 5% laked horse blood and 1% hemin and vitamin K | Anaerobic, 48 h, 37 °C |
| Peak number | RT (min) | UV absorption (λmax) | m/z [M-H] | Fragments (m/z) | Proposed Structure |
|---|---|---|---|---|---|
| 1 | 31.8 | 256 | 301 | 151, 179, 257, 273 | Quercetin |
| 2 | 35.2 | 325 | 271 | 151, 165, 225, 253 | Pinobaskin |
| 3 | 35.5 | 267, 338 | 269 | 117, 149, 225 | Apigenin |
| 4 | 44.7 | 270 | 253 | 209 | Chrysin |
| 5 | 45.5 | 290 | 255 | 151, 187, 213 | Pinocembri |
| 6 | 46.2 | 261, 351 | 269 | 227 | Galangin |
| Polyphenols | Eu1 | Eu2 | Eu3 |
|---|---|---|---|
| 1-Quercetin | 1.4 ± 0.6 | 0.8 ± 0.2 | 0.7 ± 0.4 |
| 2-Pinobanksin | 1.5 ± 0.1 | 1.0 ± 0.2 | 1.5 ± 0.3 |
| 3-Apigenin | 1.6 ± 0.3 | 1.1 ± 0.3 | 1.2 ± 0.3 |
| 4-Chrysin | 18.3 ± 0.3 | 21.4 ± 0.2 | 24.1 ± 0.4 |
| 5-Pinocembrin | 2.8 ± 0.3 | 4.8 ± 0.1 | 3.1 ± 0.2 |
| 6-Galangin | 12.6 ± 0.1 | 12.0 ± 0.1 | 12.0 ± 0.2 |
| Sum of percentages | 38.2 | 41.1 | 42.6 |
| - | Am1 | Am2 | Am3 |
| 1-Quercetin | 0.5 ± 0-6 | 0.5 ± 0.1 | 0.9 ± 0.5 |
| 2-Pinobanksin | 1.0 ± 0.2 | 0.9 ± 0.2 | 3.0 ± 0.8 |
| 3-Apigenin | 1.5 ± 0.9 | 0.9 ± 0.3 | 3.5 ± 1.2 |
| 4-Chrysin | 30.3 ± 3.3 | 22.2 ± 1.1 | 28.6 ± 0.6 |
| 5-Pinocembrin | 4.4 ± 0.4 | 1.8 ± 0.3 | 13.9 ± 1.1 |
| 6-Galangin | 15.4 ± 0.6 | 11.5 ± 0.4 | 9.4 ± 1.3 |
| Sum of percentages | 53.1 | 37.8 | 59.3 |
| - | As1 | As2 | As3 |
| 1-Quercetin | 0.4 ± 0.4 | 0.4 ± 0.5 | 0.9 ± 0.4 |
| 2-Pinobanksin | 1.0 ± 0.2 | 1.8 ± 0.4 | 10.0 ± 2.0 |
| 3-Apigenin | 2.2 ± 0.8 | 2.0 ± 1.7 | 1.2 ± 0.1 |
| 4-Chrysin | 25.0 ± 2.5 | 24.4 ± 0.8 | 19.6 ± 1.4 |
| 5-Pinocembrin | 2.0 ± 0.1 | 2.4 ± 0.1 | 1.7 ± 0.2 |
| 6-Galangin | 16.1 ± 0.6 | 15.9 ± 0.5 | 12.0 ± 1.0 |
| Sum of percentages | 46.7 | 46.9 | 45.4 |
| Comparisons | Significance | |||||
|---|---|---|---|---|---|---|
| Quercetin | Pinobanksin | Apigenin | Chrysin | Pinocembrin | Galangin | |
| EU 1 vs EU 2 | Yes * | No ** | No | No | Yes | No |
| EU 1 vs EU 3 | Yes | No | No | Yes | No | No |
| EU 1 vs AM 1 | Yes | No | No | Yes | Yes | Yes |
| EU 1 vs AM 2 | Yes | No | No | No | No | No |
| EU 1 vs AM 3 | No | No | Yes | Yes | Yes | Yes |
| EU 1 vs AS 1 | Yes | No | No | Yes | No | Yes |
| EU 1 vs AS 2 | Yes | No | No | Yes | No | Yes |
| EU 1 vs AS 3 | No | Yes | No | No | No | No |
| EU 2 vs EU 3 | No | No | No | No | Yes | No |
| EU 2 vs AM 1 | No | No | No | Yes | No | Yes |
| EU 2 vs AM 2 | No | No | No | No | Yes | No |
| EU 2 vs AM 3 | No | No | Yes | Yes | Yes | Yes |
| EU 2 vs AS 1 | No | No | Yes | No | Yes | Yes |
| EU 2 vs AS 2 | No | No | Yes | No | Yes | Yes |
| EU 2 vs AS 3 | No | Yes | No | No | Yes | No |
| EU 3 vs AM 1 | No | No | No | Yes | Yes | Yes |
| EU 3 vs AM 2 | No | No | No | No | Yes | No |
| EU 3 vs AM 3 | No | No | Yes | Yes | Yes | Yes |
| EU 3 vs AS 1 | No | No | Yes | No | No | Yes |
| EU 3 vs AS 2 | No | No | No | No | No | Yes |
| EU 3 vs AS 3 | No | Yes | No | Yes | Yes | No |
| AM 1 vs AM 2 | No | No | No | Yes | Yes | Yes |
| AM 1 vs AM 3 | No | No | Yes | No | Yes | Yes |
| AM 1 vs AS 1 | No | No | No | Yes | Yes | No |
| AM 1 vs AS 2 | No | No | No | Yes | Yes | No |
| AM 1 vs AS 3 | No | Yes | No | Yes | Yes | Yes |
| AM 2 vs AM 3 | No | No | Yes | Yes | Yes | Yes |
| AM 2 vs AS 1 | No | No | Yes | No | No | Yes |
| AM 2 vs AS 2 | No | No | Yes | No | No | Yes |
| AM 2 vs AS 3 | No | Yes | No | No | No | No |
| AM 3 vs AS 1 | No | No | Yes | No | Yes | Yes |
| AM 3 vs AS 2 | No | No | Yes | No | Yes | Yes |
| AM 3 vs AS 3 | No | Yes | Yes | Yes | Yes | Yes |
| AS 1 vs AS 2 | No | No | No | No | No | No |
| AS 1 vs AS 3 | No | Yes | Yes | Yes | No | Yes |
| AS 2 vs AS 3 | No | Yes | No | Yes | No | Yes |
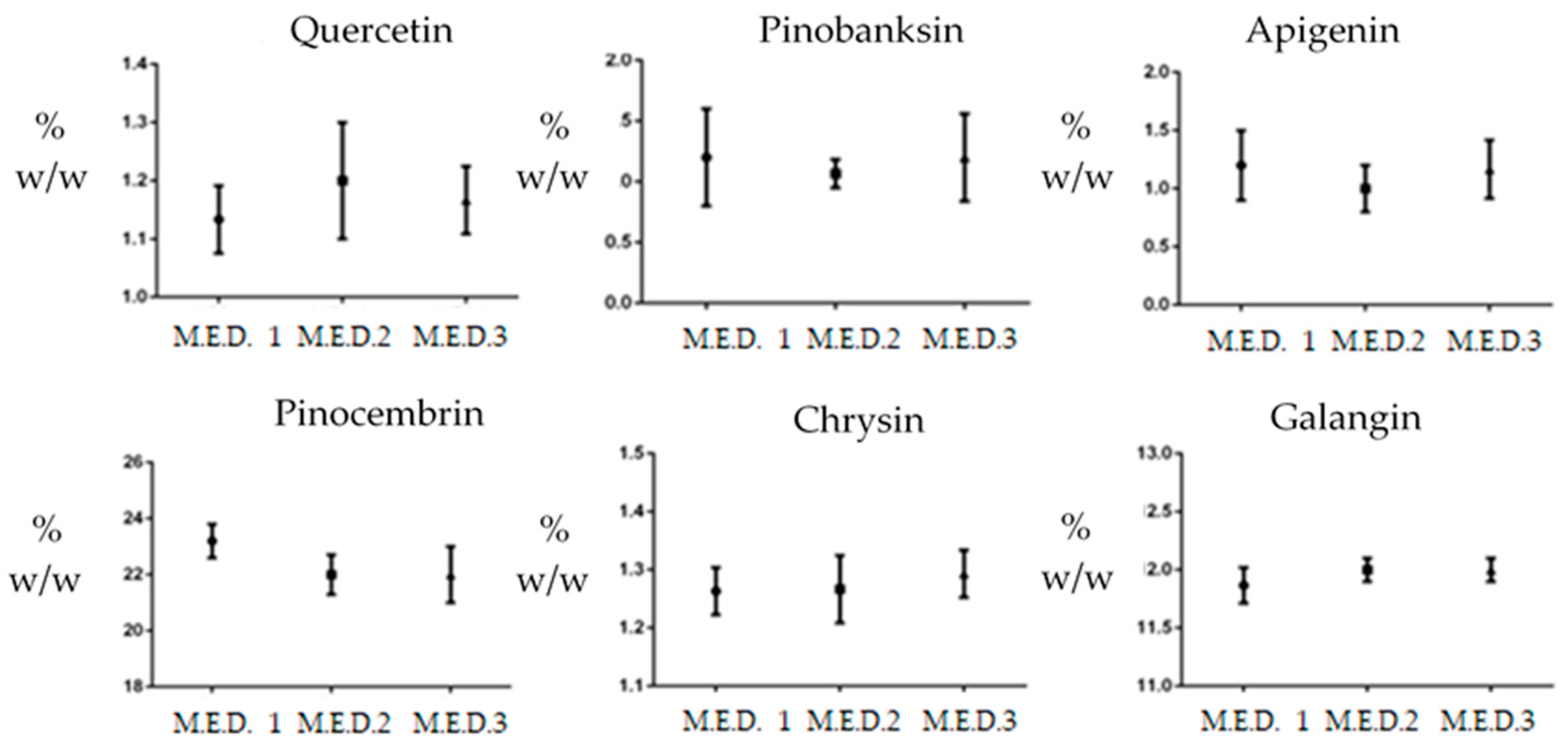
| Polyphenols | M.E.D. Propolis A | M.E.D. Propolis B | M.E.D. Propolis C |
|---|---|---|---|
| 1-Quercetin | 1.1 ± 0.05 | 1.2 ± 0.10 | 0.9 ± 0.06 |
| 2-Pinobanksin | 1.2 ± 0.40 | 0.8 ± 0.11 | 1.6 ± 0.36 |
| 3-Apigenin | 1.2 ± 0.30 | 1.0 ± 0.20 | 1.4 ± 0.04 |
| 4-Chrysin | 23.2 ± 0.60 | 22.0 ± 0.71 | 22.0 ± 1.02 |
| 5-Pinocembrin | 1.17 ± 0.04 | 1.4 ± 0.06 | 1.4 ± 0.04 |
| 6-Galangin | 13.4 ± 0.15 | 14.7 ± 0.11 | 14.3 ± 0.10 |
| Microbial Strain | MIC (µg/mL) | MIC (µg/mL) | |||
|---|---|---|---|---|---|
| CODE | A | B | C | Antimicrobial agent | |
| Staphylococcus aureus MSSA ATCC25923 | L1280 | 312 | 312 | 312 | - |
| Staphylococcus epidermidis ATCC12228 | L147 | 312 | 312 | 312 | - |
| Escherichia coli hyperpermeable | G1640 | 312 | 625 | 625 | 0.078, trimethroprim |
| Moraxella catarrhalis | L3292 | 39 | 78 | 78 | 0.3, ampicillin |
| Streptococcus pneumoniae penicillin-susceptible | L44 | 20 | 39 | 39 | 2.0, ampicillin |
| Candida albicans ATCC24443 | L4120 | 1250 | 1250 | 1250 | 0.75, fluconazole |
| Candida albicans ATCC90028 | L3023 | 1250 | 2500 | 2500 | 1.0, fluconazole |
| Candida parapsilosis ATCC90018 | L3022 | 2500 | 2500 | 2500 | 4.0, fluconazole |
| Candida kruzei | L2280 | 2500 | 2500 | 2500 | 10.0, fluconazole |
| Aspergillus niger ATCC10535 | L53 | 78 | 156 | 156 | 1000, fluconazole |
| Bacteroides fragilis ATCC25285 | L1011 | 5000 | >5000 | >5000 | 6.0, cefoxitin |
| Propionebacterium acnes ATCC25746 | L1016 | >5000 | >5000 | >5000 | 1.8, clindamycin |
| Clostridium difficile | L1365 | 2500 | 2500 | 2500 | 0.6, vancomycin |
| Clostridium difficile ATCC17858 | L4013 | 5000 | 2500 | 2500 | 1.6, vancomycin |
| Atopobium vaginae | ND736 | 156 | 156 | 156 | 0.478, ampicillin |
| Lactobacillus gasseri | ND787 | 5000 | >5000 | >5000 | 0.25, ampicillin |
| Lactobacillus acidophilus | ND786 | >5000 | >5000 | >5000 | 1.0, clindamycin |
| Neisseria gonorrhoeae | L1600 | 156 | 156 | 156 | 16.0, ampicillin |
| Neisseria gonorrhoeae | L1601 | 156 | 78 | 78 | - |
| Gardnerella vaginalis | L1629 | 312 | 312 | 156 | 0.020 ampicillin |
| Gardnerella vaginalis | L1630 | 312 | 312 | 312 | - |
| Microbial Strain | CODE | Propolis Extract A MIC (µg/mL) | Antimicrobial Agent MIC (µg/mL) |
|---|---|---|---|
| Escherichia coli | L4242 | 312 | 0.12, ampicillin |
| Staphylococcus aureus GISA MSSA | L3797 | 625 | - |
| Staphylococcus aureus GISA MRSA | L3798 | 312 | - |
| Staphylococcus haemolyticus MRSA | L1730 | 312 | - |
| Staphylococcus hominis ATCC27844 | L323 | 625 | 0.046, ampicillin |
| Staphylococcus capitis MRSA | ND021008 | 156 | - |
| Staphylococcus xylosus MRSA | ND026108 | 625 | - |
| Staphylococcus simulans | ND029808 | 1250 | - |
| Staphylococcus haemolyticus MRSA | ND040809 | 625 | - |
| Staphylococcus haemolyticus MRSA | L1729 | 1250 | - |
| Staphylococcus aureus Clinda-inducible erm(A)+ | ND053410 | 625 | 64.0, ampicillin |
| Staphylococcus aureus Community Acquired USA300 MRSA | ND054910 | 625 | - |
| Staphylococcus aureus MRSA macrolide-resistant | ND060411 | 625 | - |
| Staphylococcus aureus MRSA | L4064 | 625 | - |
| Staphylococcus epidermidis teicoplanin-resistant | ND042409 | 625 | - |
| Staphylococcus epidermidis | ND052110 | 312 | - |
| Staphylococcus epidermidis | ND051710 | 625 | - |
| Streptococcus pneumonia clindamycin/erythromycin resistant | L1542 | 39 | - |
| Streptococcus pneumonia macrolide/erythromycin resistant | L1402 | 39 | - |
| Streptococcus pneumoniae penicillin-resistant | L3917 | 20 | - |
| Candida guillermondii ATCC 6260 | L2065 | 2500 | 2.5, fluconazole |
| Candida parapsilosis ATCC22019 | L4119 | 1250 | 4.0, fluconazole |
| Escherichia coli ATCC25922 | L1281 | 5000 | 5.0, ampicillin |
| Escherichia coli | L47 | 5000 | 0.12, ampicillin |
| Pseudomonas aeruginosa ATCC27853 | L1367 | 5000 | 128.0, ampicillin |
| Acinetobacter baumannii | L3030 | 5000 | 4.0, ciprofloxacin |
| Clostridium difficile | L1366 | 5000 | 1.6, vancomycin |
| Atopobium vaginae | ND737 | 156 | 0.478, ampicillin |
| Lactobacillus paracasei | L1693 | 5000 | 0.12, penicillin |
| Lactobacillus plantarum | L19 | 5000 | 0.5, amoxicillin |
| Neisseria gonorrhoeae | L1599 | 156 | 16.0, ampicillin |
| Gardnerella vaginalis | L1622 | 312 | 0.020, ampicillin |
| Listeria monocytogenes ATCC13932 | L1450 | 1250 | 0.563, ampicillin |
| Bacillus cereus ATCC10702 | L85 | 312 | 2.0, penicillin |
| Clostridium perfringens HSR | L4053 | 5000 | 4.06, clindamycin |
| Clostridium perfringens ATCC13124 | L3697 | 2500 | 0.188, clindamycin |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zaccaria, V.; Garzarella, E.U.; Di Giovanni, C.; Galeotti, F.; Gisone, L.; Campoccia, D.; Volpi, N.; Arciola, C.R.; Daglia, M. Multi Dynamic Extraction: An Innovative Method to Obtain a Standardized Chemically and Biologically Reproducible Polyphenol Extract from Poplar-Type Propolis to Be Used for Its Anti-Infective Properties. Materials 2019, 12, 3746. https://doi.org/10.3390/ma12223746
Zaccaria V, Garzarella EU, Di Giovanni C, Galeotti F, Gisone L, Campoccia D, Volpi N, Arciola CR, Daglia M. Multi Dynamic Extraction: An Innovative Method to Obtain a Standardized Chemically and Biologically Reproducible Polyphenol Extract from Poplar-Type Propolis to Be Used for Its Anti-Infective Properties. Materials. 2019; 12(22):3746. https://doi.org/10.3390/ma12223746
Chicago/Turabian StyleZaccaria, Vincenzo, Emanuele Ugo Garzarella, Carmen Di Giovanni, Fabio Galeotti, Lucia Gisone, Davide Campoccia, Nicola Volpi, Carla Renata Arciola, and Maria Daglia. 2019. "Multi Dynamic Extraction: An Innovative Method to Obtain a Standardized Chemically and Biologically Reproducible Polyphenol Extract from Poplar-Type Propolis to Be Used for Its Anti-Infective Properties" Materials 12, no. 22: 3746. https://doi.org/10.3390/ma12223746






