Steered Pull Simulation to Determine Nanomechanical Properties of Cellulose Nanofiber
Abstract
:1. Introduction
2. Computational Methodology
2.1. Simulation Model of Cellulose Nanofiber (CNF)
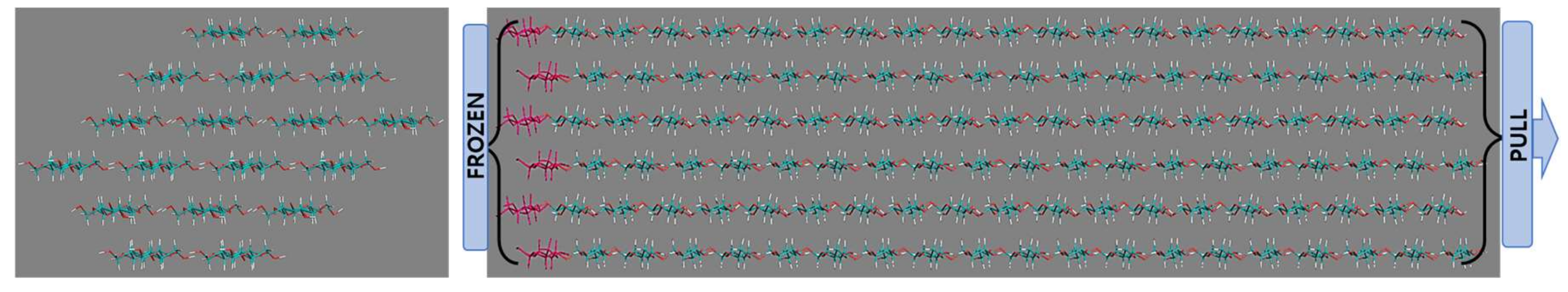
2.2. Simulation Description
3. Results and Discussion
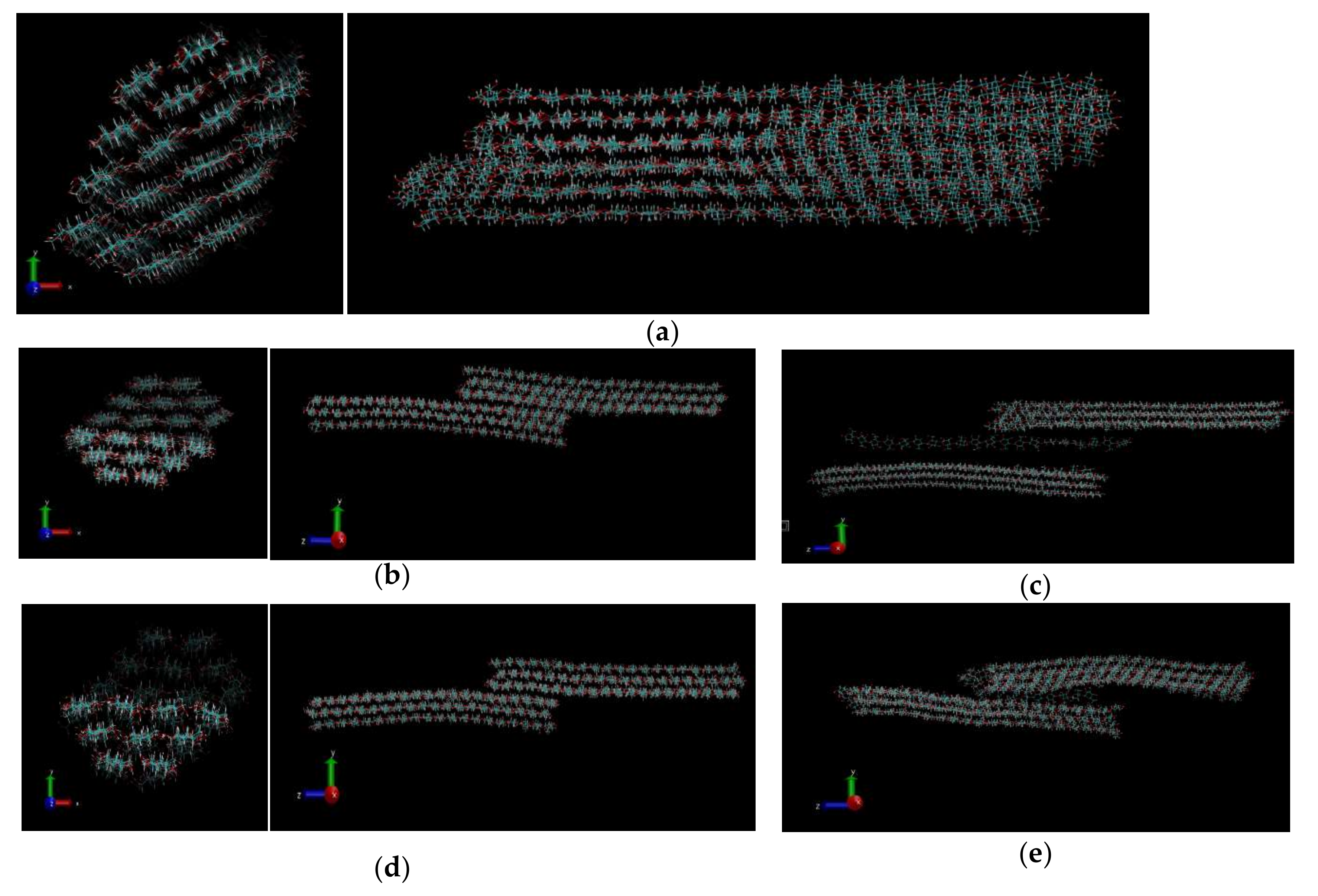
3.1. CNF Structure after Simulation
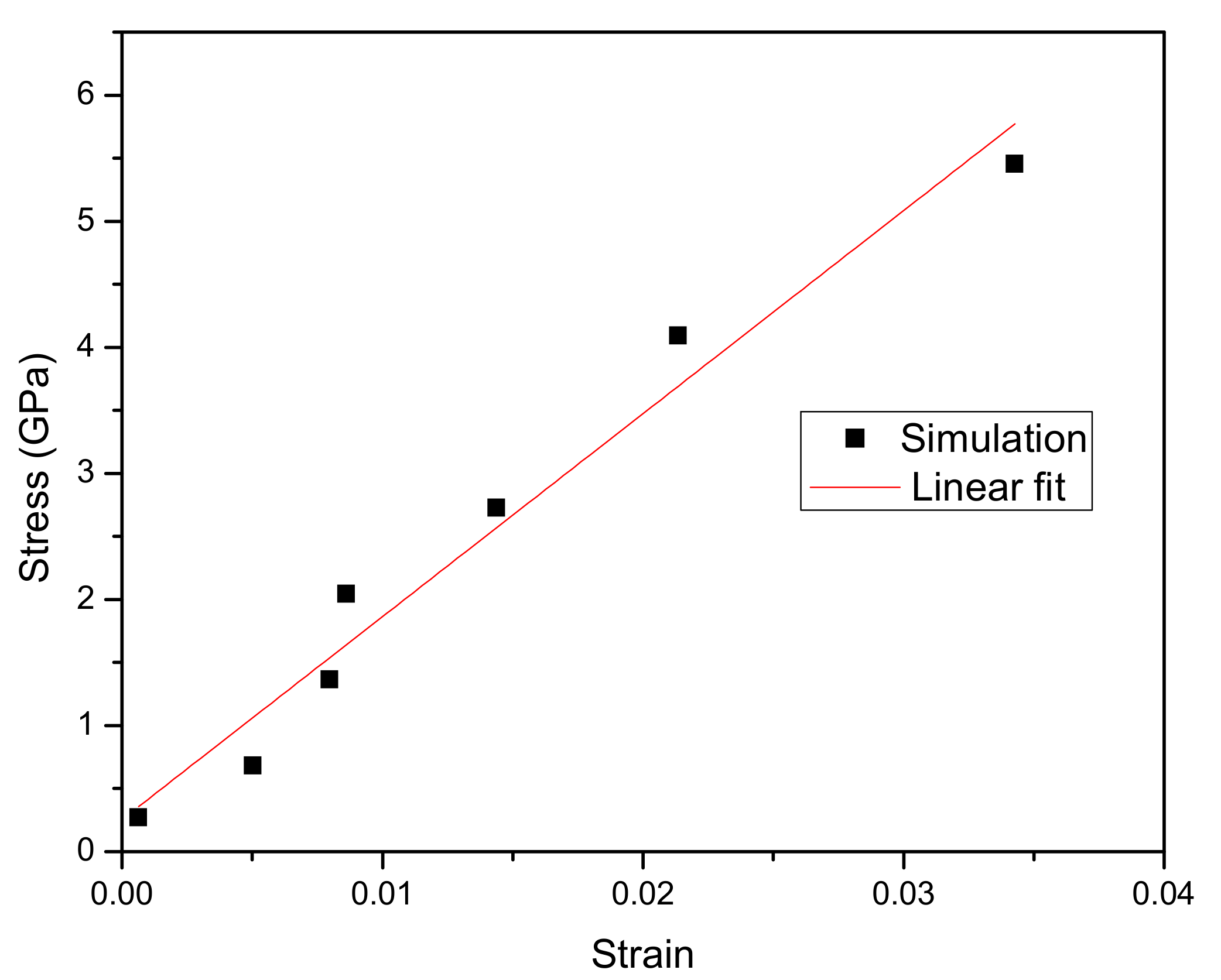
3.2. Strain Response
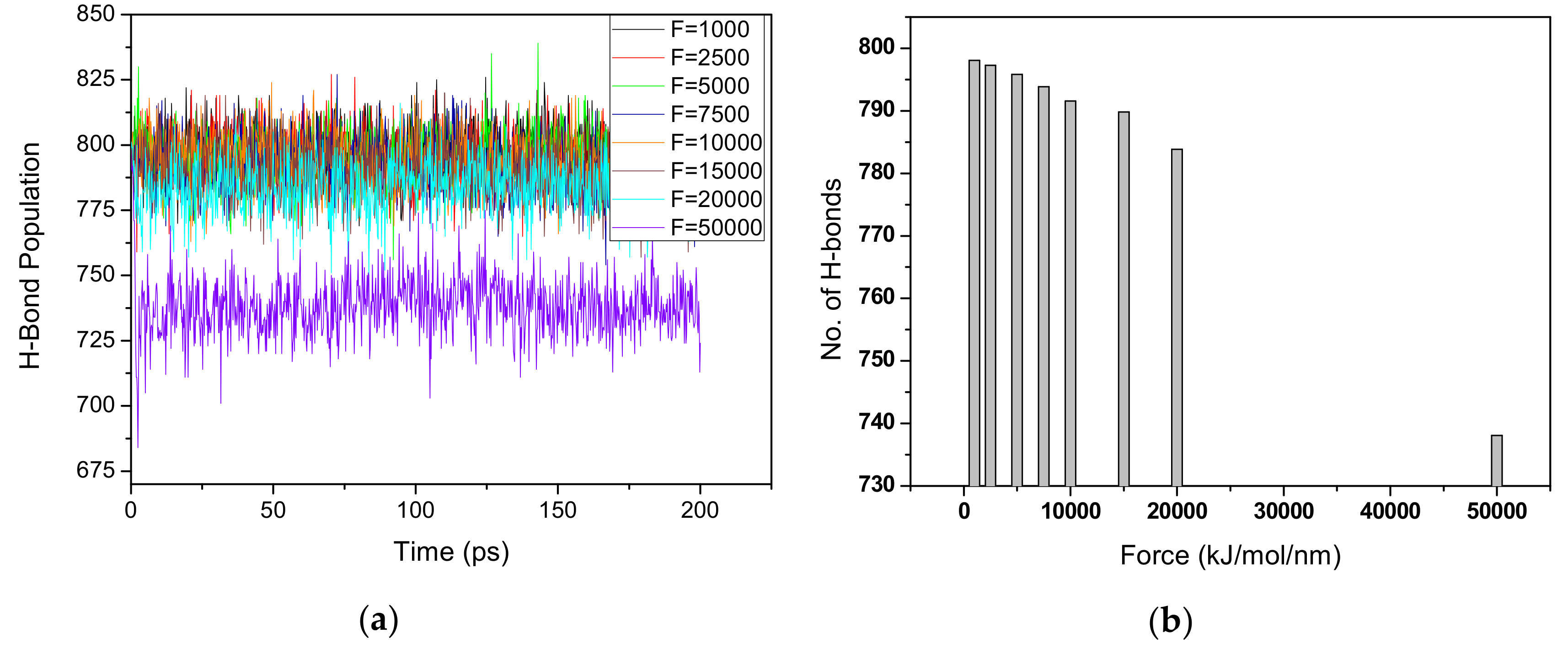
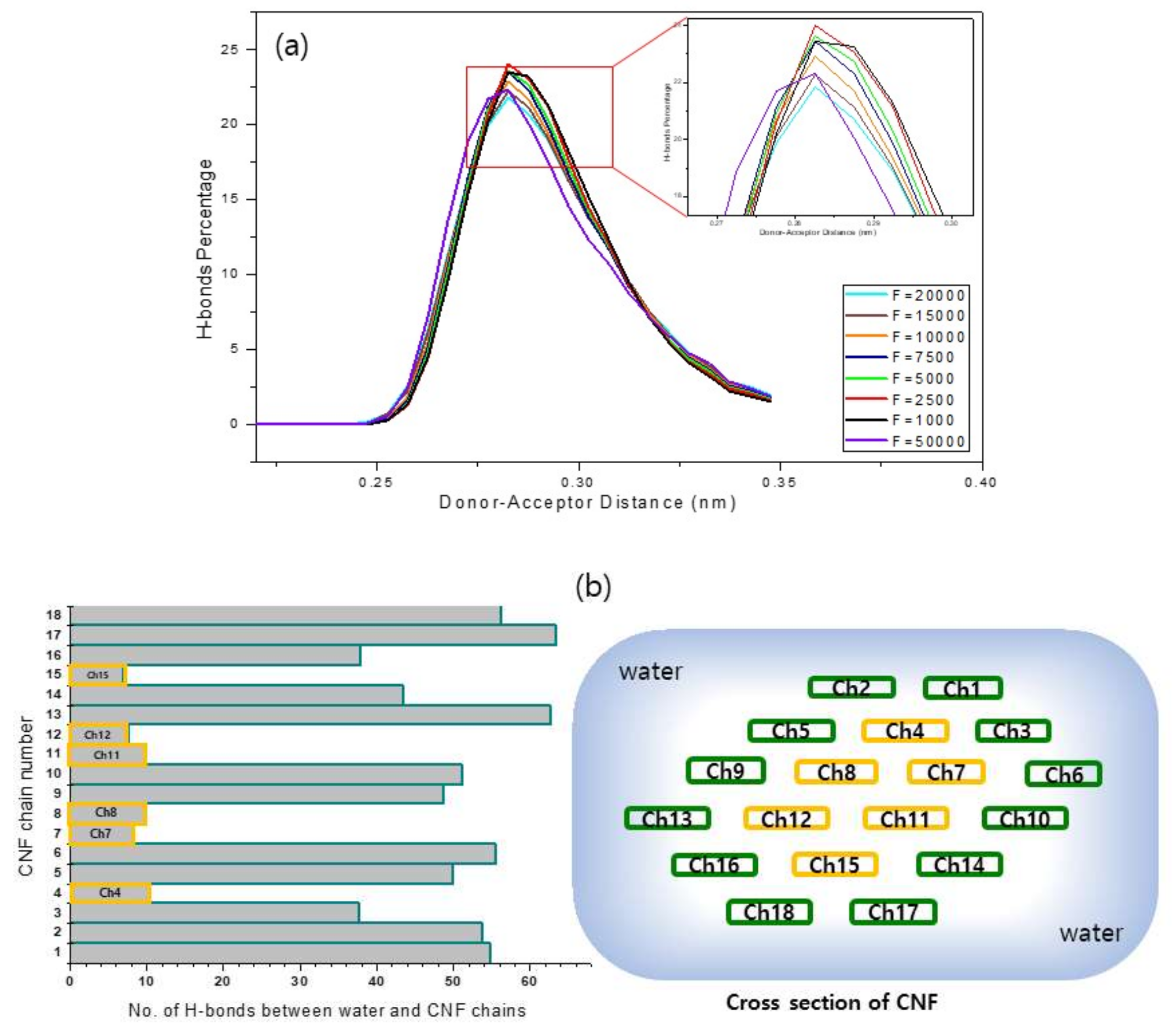
3.3. Disparity of Hydrogen Bonds
3.4. Donor-Acceptor Distance
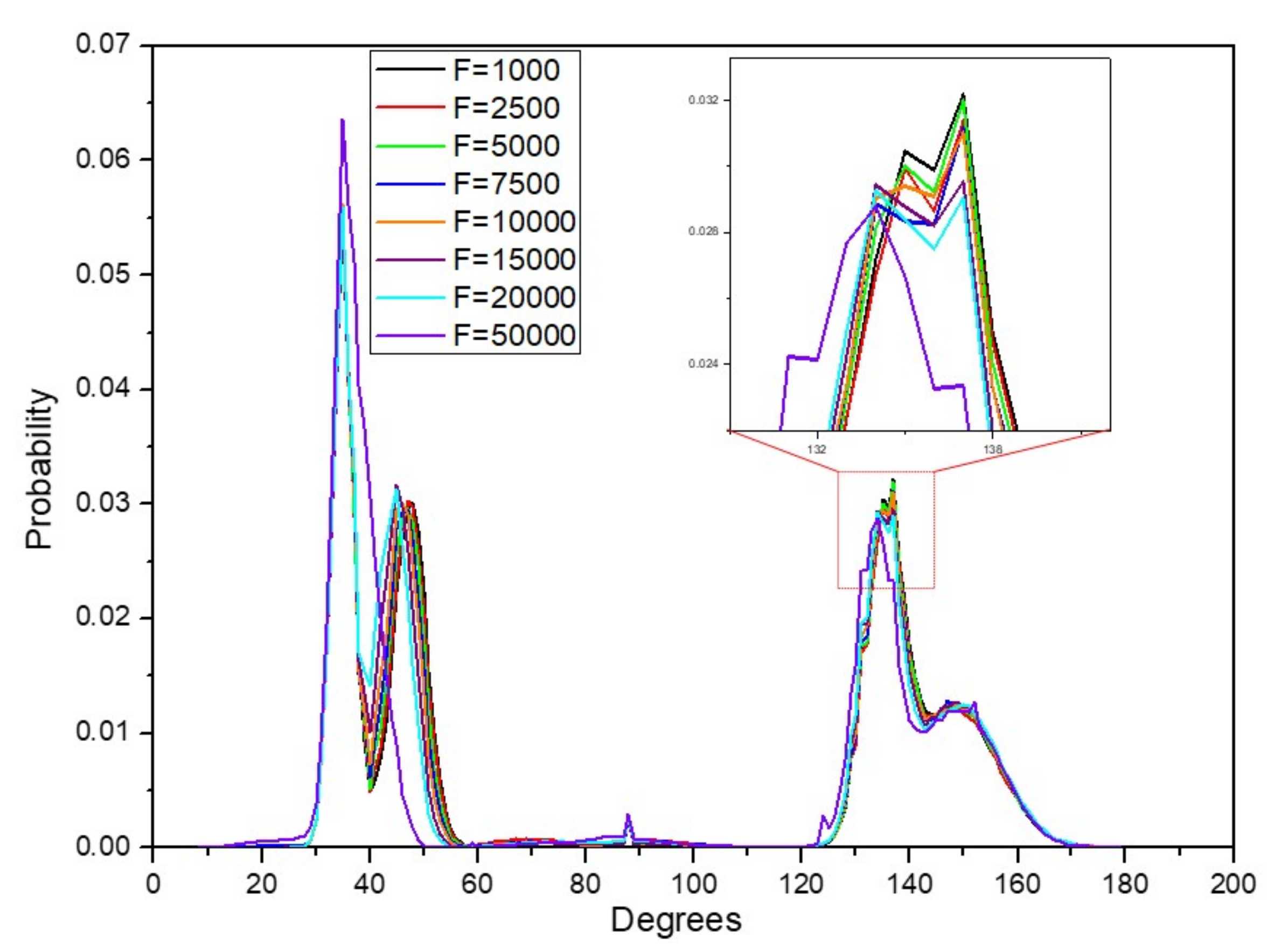
3.5. Dihedral Distribution
3.6. Mechanical Properties
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Guimarães, M.; Botaro, V.R.; Novack, K.M.; Neto, F.; Pires, W.; Mendes, L.M.; Tonoli, G.H. Preparation of cellulose nanofibrils from bamboo pulp by mechanical defibrillation for their applications in biodegradable composites. J. Nanosci. Nanotechnol. 2015, 15, 6751–6768. [Google Scholar] [CrossRef] [PubMed]
- Mondal, S. Preparation, properties and applications of nanocellulosic materials. Carbohydr. Polym. 2017, 163, 301–316. [Google Scholar] [CrossRef] [PubMed]
- Khalil, H.A.; Davoudpour, Y.; Islam, M.N.; Mustapha, A.; Sudesh, K.; Dungani, R.; Jawaid, M. Production and modification of nanofibrillated cellulose using various mechanical processes: A review. Carbohydr. Polym. 2014, 99, 649–665. [Google Scholar] [CrossRef] [PubMed]
- Schmied, F.J.; Teichert, C.; Kappel, L.; Hirn, U.; Bauer, W.; Schennach, R. What holds paper together: Nanometre scale exploration of bonding between paper fibres. Sci. Rep. 2013, 3, 2432. [Google Scholar] [CrossRef] [Green Version]
- Djahedi, C. Deformation of cellulose allomorphs studied by molecular dynamics. Ph.D. Thesis, KTH Royal Institute of Technology, Stockholm, Sweden, 18 May 2015. [Google Scholar]
- Alqus, R.W. Characterisation of Carbohydrate-graphene Interactions Using Molecular Simulation. Ph.D. Thesis, University of Manchester, Manchester, UK, 2017. [Google Scholar]
- Kroon-Batenburg, L.M.J.; Bouma, B.; Kroon, J. Stability of cellulose structures studied by MD simulations. Could mercerized cellulose II be parallel. Macromolecules 1996, 29, 5695–5699. [Google Scholar] [CrossRef]
- Bergenstråhle, M. Crystalline cellulose in bulk and at interfaces as studied by atomistic computer simulations. Ph.D. Thesis, KTH School of Chemical Science and Engineering, Stockholm, Sweden, 17 November 2008. [Google Scholar]
- Bergenstråhle, M.; Wohlert, J.; Himmel, M.E.; Brady, J.W. Simulation studies of the insolubility of cellulose. Carbohydr. Res. 2010, 345, 2060–2066. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Maxwell, D.S.; Tirado-Rives, J. Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J. Am. Chem. Soc. 1996, 118, 11225–11236. [Google Scholar] [CrossRef]
- Marhöfer, R.J.; Reiling, S.; Brickmann, J. Computer simulations of crystal structures and elastic properties of cellulose. Berichte der Bunsengesellschaft für physikalische Chemie 1996, 100, 1350–1354. [Google Scholar] [CrossRef]
- Ganister, J.; Blackwell, J. NpH-MD-simulations of the elastic moduli of cellulose II at room temperature. J. Mol. Model. 1996, 2, 278–285. [Google Scholar] [CrossRef]
- Tanaka, F.; Iwata, T. Estimation of the elastic modulus of cellulose crystal by molecular mechanics simulation. Cellulose. 2006, 1, 509–517. [Google Scholar] [CrossRef]
- Eichhorn, S.J.; Davies, G.R. Modelling the crystalline deformation of native and regenerated cellulose. Cellulose. 2006, 13, 291–307. [Google Scholar] [CrossRef]
- Bergenstråhle, M.; Berglund, L.A.; Mazeau, K. Thermal response in crystalline Iβ cellulose: A molecular dynamics study. J. Phys. Chem. B. 2007, 111, 9138–9145. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Moon, R.; Martini, A. Calculation of single chain cellulose elasticity using fully atomistic modeling. Tappi. J. 2011, 10, 37–42. [Google Scholar] [CrossRef]
- Paavilainen, S.; McWhirter, J.L.; Róg, T.; Järvinen, J.; Vattulainen, I.; Ketoja, J. Mechanical properties of cellulose nanofibers determined through atomistic molecular dynamics simulations. Nordic Pulp & Paper Res. J. 2012, 27, 282–286. [Google Scholar] [CrossRef]
- Wohlert, J.; Bergenstråhle-Wohlert, M.; Berglund, L.A. Deformation of cellulose nanocrystals: Entropy, internal energy and temperature dependence. Cellulose. 2012, 19, 1821–1836. [Google Scholar] [CrossRef]
- Wu, X.; Moon, R.J.; Martini, A. Crystalline cellulose elastic modulus predicted by atomistic models of uniform deformation and nanoscale indentation. Cellulose. 2013, 20, 43–55. [Google Scholar] [CrossRef]
- Wu, X.; Moon, R.J.; Martini, A. Tensile strength of Iβ crystalline cellulose predicted by molecular dynamics simulation. Cellulose. 2014, 21, 2233–2245. [Google Scholar] [CrossRef]
- Nishiyama, Y.; Langan, P.; Chanzy, H. Crystal structure and hydrogen-bonding system in cellulose Iβ from synchrotron X-ray and neutron fiber diffraction. J. Am. Chem. Soc. 2002, 124, 9074–9082. [Google Scholar] [CrossRef]
- Ding, S.Y.; Zhao, S.; Zeng, Y. Size, shape, and arrangement of native cellulose fibrils in maize cell walls. Cellulose 2014, 21, 863–871. [Google Scholar] [CrossRef]
- Gomes, T.C.; Skaf, M.S. Cellulose-Builder: A toolkit for building crystalline structures of cellulose. J. Comput. Chem. 2012, 33, 1338–1346. [Google Scholar] [CrossRef] [PubMed]
- Thomas, L.H.; Forsyth, V.T.; Martel, A.; Grillo, I.; Altaner, C.M.; Jarvis, M.C. Structure and spacing of cellulose microfibrils in woody cell walls of dicots. Cellulose 2014, 21, 3887–3895. [Google Scholar] [CrossRef]
- Thomas, L.H.; Forsyth, V.T.; Martel, A.; Grillo, I.; Altaner, C.M.; Jarvis, M.C. Diffraction evidence for the structure of cellulose microfibrils in bamboo, a model for grass and cereal celluloses. BMC Plant Biol. 2015, 15, 153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomas, L.H.; Forsyth, V.T.; Šturcová, A.; Kennedy, C.J.; May, R.P.; Altaner, C.M.; Apperley, D.C.; Wess, T.J.; Jarvis, M.C. Structure of cellulose microfibrils in primary cell walls from collenchyma. Plant Physiol. 2013, 161, 465–476. [Google Scholar] [CrossRef] [Green Version]
- Lundborg, M.; Lindahl, E. Automatic GROMACS topology generation and comparisons of force fields for solvation free energy calculations. J. Phy. Chem. B. 2015, 119, 810–823. [Google Scholar] [CrossRef] [Green Version]
- Damm, W.; Frontera, A.; Tirado–Rives, J.; Jorgensen, W.L. OPLS all-atom force field for carbohydrates. J. Comput. Chem. 1997, 18, 1955–1970. [Google Scholar] [CrossRef]
- Kaminski, G.A.; Friesner, R.A.; Tirado-Rives, J.; Jorgensen, W.L. Evaluation and reparametrization of the OPLS-AA force field for proteins via comparison with accurate quantum chemical calculations on peptides. J. Phys. Chem. B 2001, 105, 6474–6487. [Google Scholar] [CrossRef]
- Kony, D.; Damm, W.; Stoll, S.; Van Gunsteren, W.F. An improved OPLS–AA force field for carbohydrates. J. Comput. Chem. 2002, 23, 1416–1429. [Google Scholar] [CrossRef]
- Price, M.L.; Ostrovsky, D.; Jorgensen, W.L. Gas-phase and liquid-state properties of esters, nitriles, and nitro compounds with the OPLS-AA force field. J. Comput. Chem. 2001, 22, 1340–1352. [Google Scholar] [CrossRef]
- Rizzo, R.C.; Jorgensen, W.L. OPLS all-atom model for amines: Resolution of the amine hydration problem. J. Am. Chem. Soc. 1999, 21, 4827–4836. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Tirado-Rives, J. The OPLS [optimized potentials for liquid simulations] potential functions for proteins, energy minimizations for crystals of cyclic peptides and crambin. J. Am. Chem. Soc. 1988, 110, 1657–1666. [Google Scholar] [CrossRef] [PubMed]
- Maurer, R.J.; Sax, A.F.; Ribitsch, V. Molecular simulation of surface reorganization and wetting in crystalline cellulose I and II. Cellulose 2013, 20, 25–42. [Google Scholar] [CrossRef] [Green Version]
- Srinivas, G.; Cheng, X.; Smith, J.C. Coarse-Grain Model for Natural Cellulose Fibrils in Explicit Water. J. Phys. Chem. B. 2014, 118, 3026–3034. [Google Scholar] [CrossRef] [PubMed]
- Hirn, U.; Schennach, R. Comprehensive analysis of individual pulp fiber bonds quantifies the mechanisms of fiber bonding in paper. Sci. Rep. 2015, 5, 10503. [Google Scholar] [CrossRef] [Green Version]
- Horii, F.; Yamamoto, H.; Kitamaru, R.; Tanahashi, M.; Higuchi, T. Transformation of native cellulose crystals induced by saturated steam at high temperatures. Macromolecules 1987, 11, 2946–2949. [Google Scholar] [CrossRef]
- Srivastava, P.K.; Venkatraman, K. Structure, molecular dynamics, and stress in a linear polymer. Mech. Mater. 2010, 61, 49–59. [Google Scholar] [CrossRef]
- Voss, N.R.; Gerstein, M. 3V: Cavity, channel and cleft volume calculator and extractor. Nucleic Acids Res. 2010, 38, W555–W562. [Google Scholar] [CrossRef] [Green Version]




© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Muthoka, R.M.; Kim, H.C.; Kim, J.W.; Zhai, L.; Panicker, P.S.; Kim, J. Steered Pull Simulation to Determine Nanomechanical Properties of Cellulose Nanofiber. Materials 2020, 13, 710. https://doi.org/10.3390/ma13030710
Muthoka RM, Kim HC, Kim JW, Zhai L, Panicker PS, Kim J. Steered Pull Simulation to Determine Nanomechanical Properties of Cellulose Nanofiber. Materials. 2020; 13(3):710. https://doi.org/10.3390/ma13030710
Chicago/Turabian StyleMuthoka, Ruth M., Hyun Chan Kim, Jung Woong Kim, Lindong Zhai, Pooja S. Panicker, and Jaehwan Kim. 2020. "Steered Pull Simulation to Determine Nanomechanical Properties of Cellulose Nanofiber" Materials 13, no. 3: 710. https://doi.org/10.3390/ma13030710




