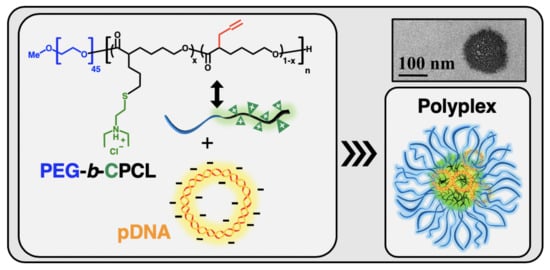
PEGylated Amine-Functionalized Poly(ε-caprolactone) for the Delivery of Plasmid DNA
Abstract
:1. Introduction
2. Materials and Methods
2.1. Measurements
2.2. Materials
2.3. Synthesis of Allyl-Functionalized CL (ACL, 1)
2.4. Synthesis of PEG-b-(Allyl-Functionalized PCL) (PEG-b-APCL, 2)
2.5. Synthesis of PEG-b-CPCL-70 (3)
2.6. Synthesis of Allyl-Functionalized Poly(ε-caprolactone) (APCL) (4)
2.7. Synthesis of Cationic Poly(ε-caprolactone) (CPCL) (5)
2.8. Study of Hydrolytic Stability of PEG-b-CPCL-70 (3)
2.9. Preparation of Polymer:pDNA Nanocomplexes
2.10. N/P Ratio Calculation
2.11. pDNA Gel-shift Assay
2.12. pDNA Release by Polyanionic Heparin
2.13. Serum Stability of Polymer:pDNA Nanocomplexes
3. Results and Discussion
3.1. Synthesis and Characterization of PEG-b-CPCL-70 (3) and CPCLs (5)
3.2. Hydrolytic Stability of PEG-b-CPCL-70 (3)
3.3. Nanocomplex Formation and Characterization
3.4. pDNA Release and Serum Stability of Nanocomplexes
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Rosenberg, S.A.; Aebersold, P.; Cornetta, K.; Kasid, A.; Morgan, R.A.; Moen, R.; Karson, E.M.; Lotze, M.T.; Yang, J.C.; Topalian, S.L. Gene transfer into humans—Immunotherapy of patients with advanced melanoma, using tumor-infiltrating lymphocytes modified by retroviral gene transduction. N. Engl. J. Med. 1990, 323, 570–578. [Google Scholar] [CrossRef] [PubMed]
- Ginn, S.L.; Amaya, A.K.; Alexander, I.E.; Edelstein, M.; Abedi, M.R. Gene therapy clinical trials worldwide to 2017: An update. J. Gene Med. 2018, 20, e3015. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Zeng, X.; Liu, M.; Deng, Y.; He, N. Current progress in gene delivery technology based on chemical methods and nano-carriers. Theranostics 2014, 4, 240–255. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Kanasty, R.L.; Eltoukhy, A.A.; Vegas, A.J.; Dorkin, J.R.; Anderson, D.G. Non-viral vectors for gene-based therapy. Nat. Rev. Genet. 2014, 15, 541–555. [Google Scholar] [CrossRef]
- Mehier-Humbert, S.; Guy, R.H. Physical methods for gene transfer: Improving the kinetics of gene delivery into cells. Adv. Drug Deliv. Rev. 2005, 57, 733–753. [Google Scholar] [CrossRef]
- Jones, C.H.; Chen, C.-K.; Ravikrishnan, A.; Rane, S.; Pfeifer, B.A. Overcoming nonviral gene delivery barriers: Perspective and future. Mol. Pharm. 2013, 10, 4082–4098. [Google Scholar] [CrossRef] [Green Version]
- Capecchi, M.R. High efficiency transformation by direct microinjection of DNA into cultured mammalian cells. Cell 1980, 22, 479–488. [Google Scholar] [CrossRef]
- Klein, T.M.; Wolf, E.D.; Wu, R.; Sanford, J.C. High-velocity microprojectiles for delivering nucleic acids into living cells. Nature 1987, 327, 70–73. [Google Scholar] [CrossRef]
- Yang, N.-S.; Burkholder, J.; Roberts, B.; Martinell, B.; McCabe, D. In vivo and in vitro gene transfer to mammalian somatic cells by particle bombardment. Proc. Natl. Acad. Sci. USA 1990, 87, 9568–9572. [Google Scholar] [CrossRef] [Green Version]
- Neumann, E.; Schaefer-Ridder, M.; Wang, Y.; Hofschneider, P.H. Gene transfer into mouse lyoma cells by electroporation in high electric fields. EMBO J. 1982, 1, 841–845. [Google Scholar] [CrossRef]
- Kim, H.J.; Greenleaf, J.F.; Kinnick, R.R.; Bronk, J.T.; Bolander, M.E. Ultrasound-mediated transfection of mammalian cells. Hum. Gene Ther. 1996, 7, 1339–1346. [Google Scholar] [CrossRef] [PubMed]
- Tao, W.; Wilkinson, J.; Stanbridge, E.J.; Berns, M.W. Direct gene transfer into human cultured cells facilitated by laser micropuncture of the cell membrane. Proc. Natl. Acad. Sci. USA 1987, 84, 4180–4184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scherer, F.; Anton, M.; Schillinger, U.; Henke, J.; Bergemann, C.; Krüger, A.; Gänsbacher, B.; Plank, C. Magnetofection: Enhancing and targeting gene delivery by magnetic force in vitro and in vivo. Gene Ther. 2002, 9, 102–109. [Google Scholar] [CrossRef] [Green Version]
- Lechardeur, D.; Sohn, K.J.; Haardt, M.; Joshi, P.B.; Monck, M.; Graham, R.W.; Beatty, B.; Squire, J.; O’Brodovich, H.; Lukacs, G.L. Metabolic instability of plasmid DNA in the cytosol: A Potential barrier to gene transfer. Gene Ther. 1999, 6, 482–497. [Google Scholar] [CrossRef] [Green Version]
- Houk, B.E.; Hochhaus, G.; Hughes, J.A. Kinetic modeling of plasmid DNA degradation in rat plasma. AAPS PharmSci 1999, 1, 15–20. [Google Scholar] [CrossRef] [Green Version]
- Bessis, N.; GarciaCozar, F.J.; Boissier, M.C. Immune responses to gene therapy vectors: Influence on vector function and effector mechanisms. Gene Ther. 2004, 11, 10–17. [Google Scholar] [CrossRef] [Green Version]
- Bouard, D.; Alazard-Dany, N.; Cosset, F.L. Viral vectors: From virology to transgene expression. Br. J. Pharmacol. 2009, 157, 153–165. [Google Scholar] [CrossRef] [Green Version]
- Shim, G.; Kim, D.; Le, Q.-V.; Park, G.T.; Kwon, T.; Oh, Y.-K. Nonviral Delivery Systems for Cancer Gene Therapy: Strategies and Challenges. Curr. Gene Ther. 2018, 18, 3–20. [Google Scholar] [CrossRef]
- Li, S.; Huang, L. Nonviral gene therapy: Promises and challenges. Gene Ther. 2000, 7, 31–34. [Google Scholar] [CrossRef] [Green Version]
- Roy, K.; Mao, H.-Q.; Huang, S.K.; Leong, K.W. Oral gene delivery with chitosan–DNA nanoparticles generates immunologic protection in a murine model of peanut allergy. Nat. Med. 1999, 5, 387–391. [Google Scholar] [CrossRef]
- Akinc, A.; Anderson, D.G.; Lynn, D.M.; Langer, R. Synthesis of poly(β-amino ester)s optimized for highly effective gene delivery. Bioconjugate Chem. 2003, 14, 979–988. [Google Scholar] [CrossRef] [PubMed]
- Lim, Y.-B.; Kim, C.-H.; Kim, K.; Kim, S.W.; Park, J.-S. Development of a safe gene delivery system using biodegradable polymer, poly [α-(4-aminobutyl)-L-glycolic acid]. J. Am. Chem. Soc. 2000, 122, 6524–6525. [Google Scholar] [CrossRef]
- Zhang, Z.; Yin, L.; Xu, Y.; Tong, R.; Lu, Y.; Ren, J.; Cheng, J. Facile functionalization of polyesters through thiol-yne chemistry for the design of degradable, cell-penetrating and gene delivery dual-functional agents. Biomacromolecules 2012, 13, 3456–3462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, H.; Zhu, D.; Zhou, Z.; Liu, X.; Piao, Y.; Zhang, Z.; Liu, X.; Tang, J.; Shen, Y. Facile synthesis of semi-library of low charge density cationic polyesters from poly(alkylene maleate)s for efficient local gene delivery. Biomaterials 2018, 178, 559–569. [Google Scholar] [CrossRef]
- Chen, C.-K.; Jones, C.H.; Mistriotis, P.; Yu, Y.; Ma, X.; Ravikrishnan, A.; Jiang, M.; Andreadis, S.T.; Pfeifer, B.A.; Cheng, C. Poly(ethylene glycol)-block-cationic polylactide nanocomplexes of differing charge density for gene delivery. Biomaterials 2013, 34, 9688–9699. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-K.; Law, W.-C.; Aalinkeel, R.; Nair, B.; Kopwitthaya, A.; Mahajan, S.D.; Reynolds, J.L.; Zou, J.; Schwartz, S.A.; Prasad, P.N.; et al. Well-Defined Degradable Cationic Polylactide as Nanocarrier for the Delivery of siRNA to Silence Angiogenesis in Prostate Cancer. Adv. Healthc. Mater. 2012, 1, 751–761. [Google Scholar] [CrossRef]
- Jones, C.H.; Chen, C.-K.; Chen, M.; Ravikrishnan, A.; Zhang, H.; Gollakota, A.; Chung, T.; Cheng, C.; Pfeifer, B.A. PEGylated cationic polylactides for hybrid biosynthetic gene delivery. Mol. Pharm. 2015, 12, 846–856. [Google Scholar] [CrossRef]
- Chen, C.-K.; Law, W.-C.; Aalinkeel, R.; Yu, Y.; Nair, B.; Wu, J.; Mahajan, S.; Reynolds, J.L.; Li, Y.; Lai, C.K.; et al. Biodegradable cationic polymeric nanocapsules for overcoming multidrug resistance and enabling drug–gene co-delivery to cancer cells. Nanoscale 2014, 6, 1567–1572. [Google Scholar] [CrossRef] [Green Version]
- Jones, C.H.; Chen, C.-K.; Jiang, M.; Fang, L.; Cheng, C.; Pfeifer, B.A. Synthesis of cationic polylactides with tunable charge densities as nanocarriers for effective gene delivery. Mol. Pharm. 2013, 10, 1138–1145. [Google Scholar] [CrossRef]
- Darcos, V.; El Habnouni, S.; Nottelet, B.; El Ghzaoui, A.; Coudane, J. Well-defined PCL-graft-PDMAEMA prepared by ring-opening polymerisation and click chemistry. Polym. Chem. 2010, 1, 280–282. [Google Scholar] [CrossRef]
- Lohmeijer, B.G.; Pratt, R.C.; Leibfarth, F.; Logan, J.W.; Long, D.A.; Dove, A.P.; Nederberg, F.; Choi, J.; Wade, C.; Waymouth, R.M. Guanidine and amidine organocatalysts for ring-opening polymerization of cyclic esters. Macromolecules 2006, 39, 8574–8583. [Google Scholar] [CrossRef]
- Pelegri-O’Day, E.M.; Paluck, S.J.; Maynard, H.D. Substituted polyesters by thiol–ene modification: Rapid diversification for therapeutic protein stabilization. J. Am. Chem. Soc. 2017, 139, 1145–1154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Felgner, P.; Barenholz, Y.; Behr, J.; Cheng, S.; Cullis, P.; Huang, L.; Jessee, J.; Seymour, L.; Szoka, F.; Thierry, A. Nomenclature for synthetic gene delivery systems. Hum. Gene Ther. 1997, 8, 511–512. [Google Scholar] [CrossRef] [PubMed]
- Perevyazko, I.Y.; Bauer, M.; Pavlov, G.M.; Hoeppener, S.; Schubert, S.; Fischer, D.; Schubert, U.S. Polyelectrolyte complexes of DNA and linear PEI: Formation, composition and properties. Langmuir 2012, 28, 16167–16176. [Google Scholar] [CrossRef]
- Xiong, X.-B.; Uludağ, H.; Lavasanifar, A. Biodegradable amphiphilic poly (ethylene oxide)-block-polyesters with grafted polyamines as supramolecular nanocarriers for efficient siRNA delivery. Biomaterials 2009, 30, 242–253. [Google Scholar] [CrossRef]
- Adami, R.C.; Collard, W.T.; Gupta, S.A.; Kwok, K.Y.; Bonadio, J.; Rice, K.G. Stability of peptide-condensed plasmid DNA formulations. J. Pharm. Sci. 1998, 87, 678–683. [Google Scholar] [CrossRef]
- Nair, L.S.; Laurencin, C.T. Biodegradable polymers as biomaterials. Prog. Polym. Sci. 2007, 32, 762–798. [Google Scholar] [CrossRef]
- Li, S.; Garreau, H.; Vert, M.; Petrova, T.; Manolova, N.; Rashkov, I. Hydrolytic degradation of poly (oxyethylene)–poly-(ε-caprolactone) multiblock copolymers. J. Appl. Polym. Sci. 1998, 68, 989–998. [Google Scholar] [CrossRef]
- Kohler, J.; Marquardt, F.; Teske, M.; Keul, H.; Sternberg, K.; Moller, M. Enhanced hydrolytic degradation of heterografted polyglycidols: Phosphonoethylated monoester and polycaprolactone grafts. Biomacromolecules 2013, 14, 3985–3996. [Google Scholar] [CrossRef]
- Agostini, A.; Gatti, S.; Cesana, A.; Moscatelli, D. Synthesis and degradation study of cationic polycaprolactone-based nanoparticles for biomedical and industrial applications. Ind. Eng. Chem. Res. 2017, 56, 5872–5880. [Google Scholar] [CrossRef]
- Jacobs, F.; Wisse, E.; De Geest, B. The role of liver sinusoidal cells in hepatocyte-directed gene transfer. Am. J. Pathol. 2010, 176, 14–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wisse, E.; Jacobs, F.; Topal, B.; Frederik, P.; De Geest, B. The size of endothelial fenestrae in human liver sinusoids: Implications for hepatocyte-directed gene transfer. Gene Ther. 2008, 15, 1193–1199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jafari, A.; Sun, H.; Sun, B.; Mohamed, M.A.; Cui, H.; Cheng, C. Layer-by-layer preparation of polyelectrolyte multilayer nanocapsules via crystallized miniemulsions. Chem. Commun. 2019, 55, 1267–1270. [Google Scholar] [CrossRef] [PubMed]
















| Polymer | Solubility in | |
|---|---|---|
| PBS pH 5.5 | PBS pH 7.4 | |
| PEG-b-CPCL-70 (3) | √ | √ |
| CPCL-50 | √ | × |
| CPCL-95 | √ | × |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jafari, A.; Rajabian, N.; Zhang, G.; Alaa Mohamed, M.; Lei, P.; Andreadis, S.T.; Pfeifer, B.A.; Cheng, C. PEGylated Amine-Functionalized Poly(ε-caprolactone) for the Delivery of Plasmid DNA. Materials 2020, 13, 898. https://doi.org/10.3390/ma13040898
Jafari A, Rajabian N, Zhang G, Alaa Mohamed M, Lei P, Andreadis ST, Pfeifer BA, Cheng C. PEGylated Amine-Functionalized Poly(ε-caprolactone) for the Delivery of Plasmid DNA. Materials. 2020; 13(4):898. https://doi.org/10.3390/ma13040898
Chicago/Turabian StyleJafari, Amin, Nika Rajabian, Guojian Zhang, Mohamed Alaa Mohamed, Pedro Lei, Stelios T. Andreadis, Blaine A. Pfeifer, and Chong Cheng. 2020. "PEGylated Amine-Functionalized Poly(ε-caprolactone) for the Delivery of Plasmid DNA" Materials 13, no. 4: 898. https://doi.org/10.3390/ma13040898