Opportunities Offered by Graphene Nanoparticles for MicroRNAs Delivery for Amyotrophic Lateral Sclerosis Treatment
Abstract
:1. Introduction
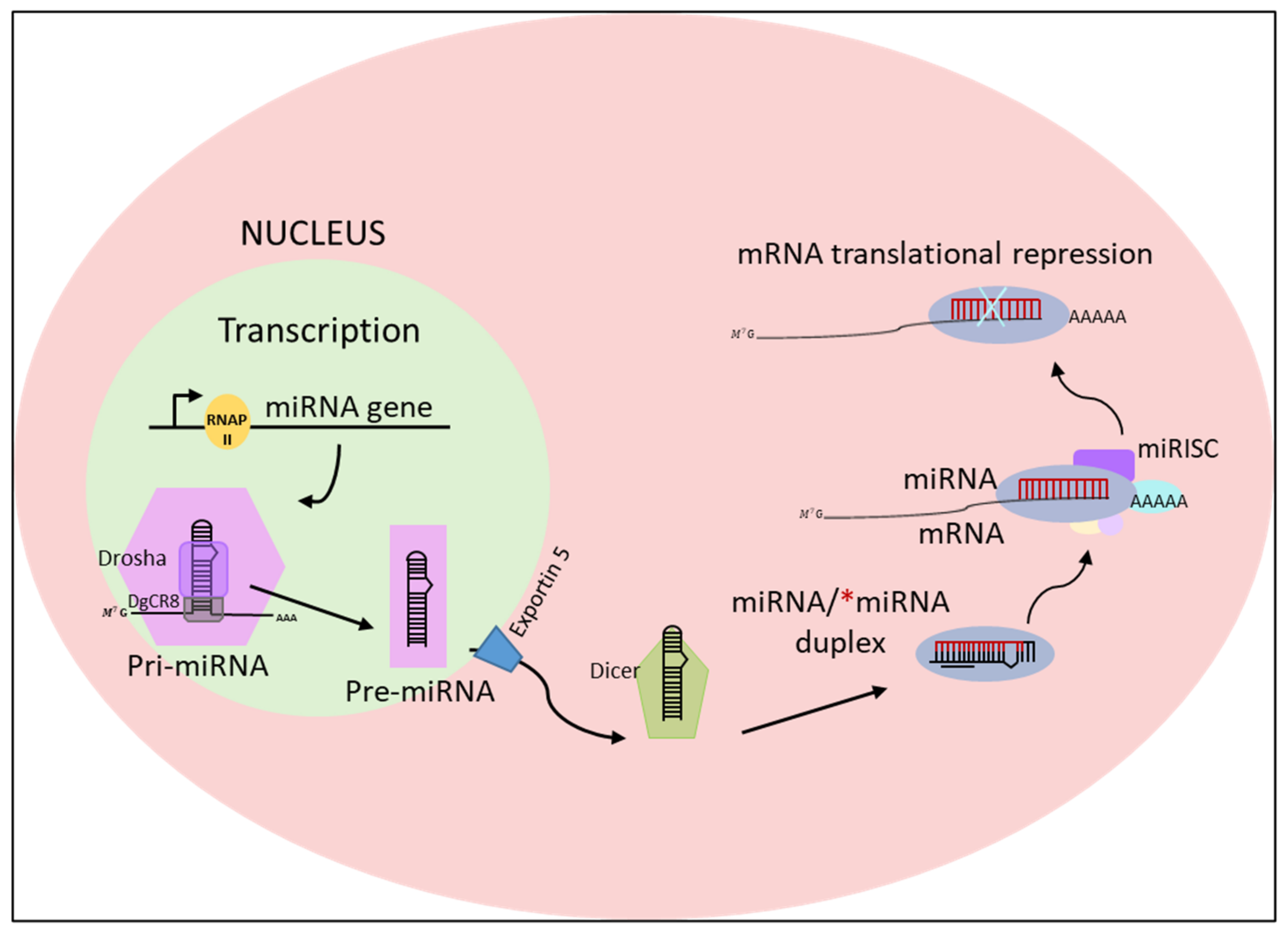
2. MicroRNAs: Biogenesis, Function, and Mechanism of Action
3. MiRNAs Dysfunction in ALS and Their Uses as Biomarkers of Disease
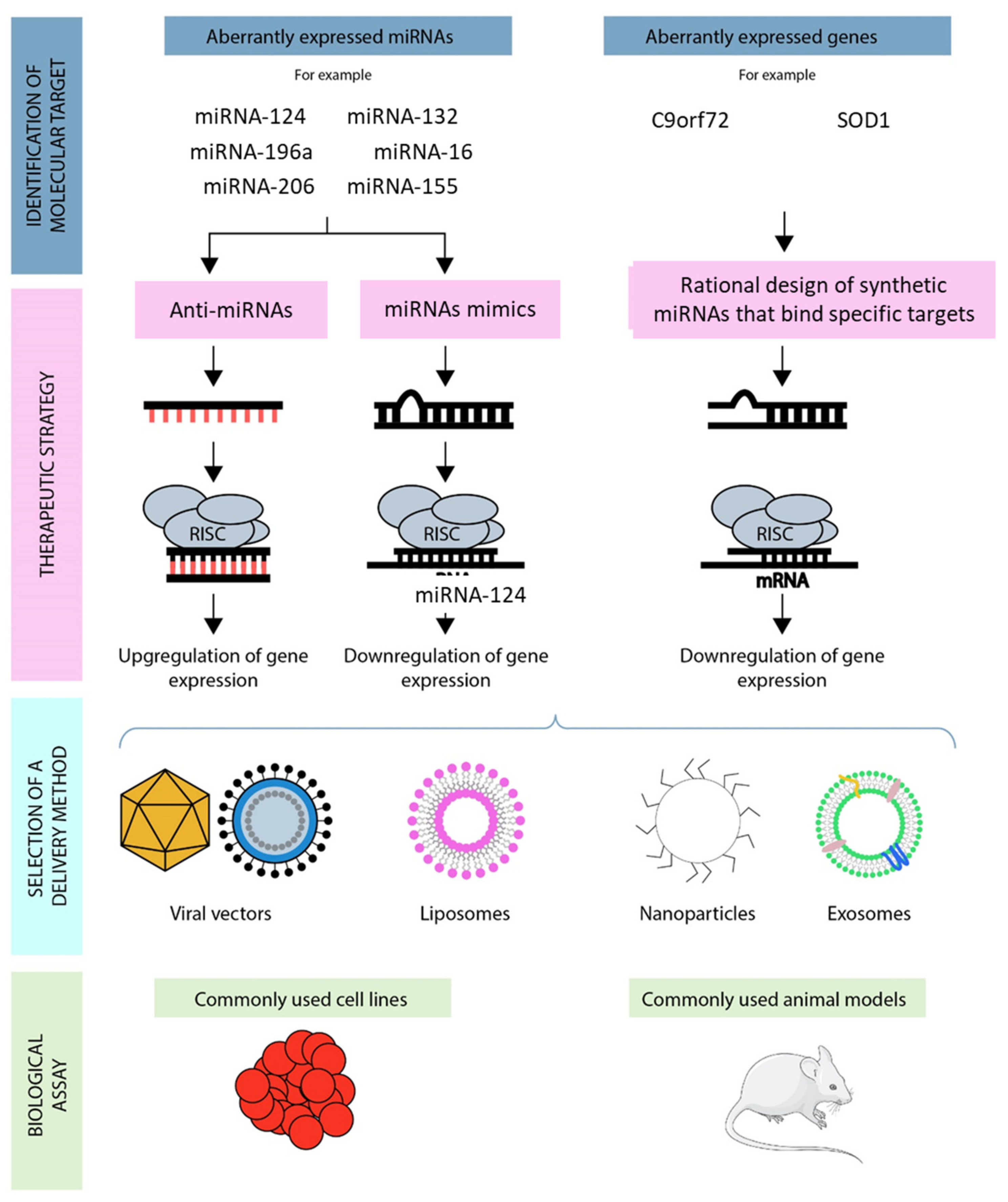
4. MicroRNAs and the Treatment of ALS
4.1. MiRNA Inhibition Therapy
4.2. MiRNA Replacement Therapy
5. Approaches for MiRNA Therapeutic Delivery
5.1. MiRNA Delivery Systems
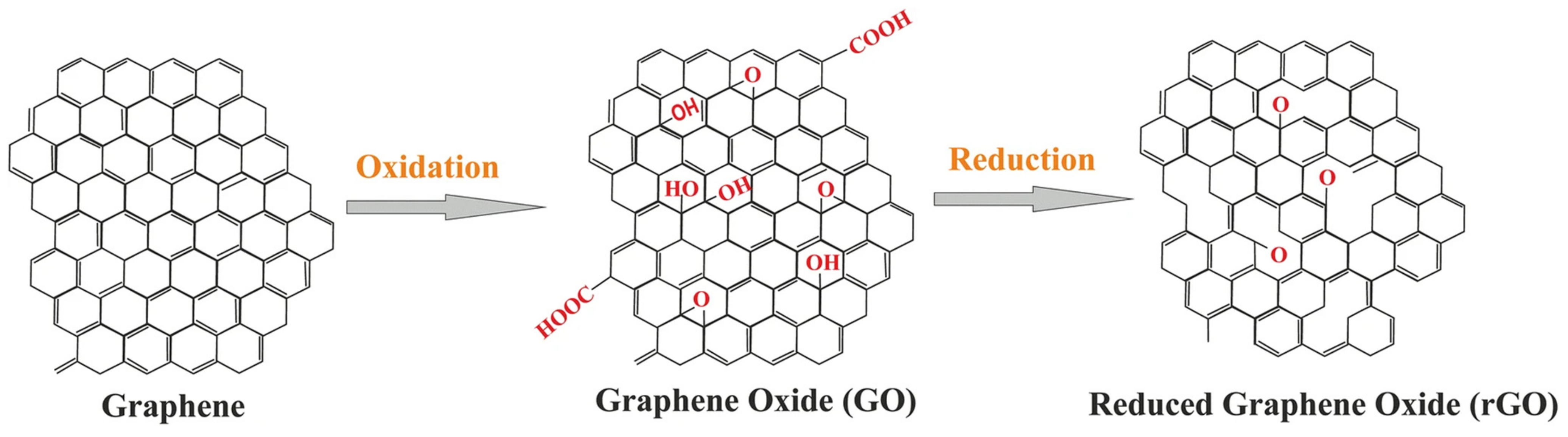
5.2. Graphene-Mediated miRNA Delivery
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Masrori, P.; Van Damme, P. Amyotrophic lateral sclerosis: A clinical review. Eur. J. Neurol. 2020, 27, 1918–1929. [Google Scholar] [CrossRef] [PubMed]
- van Es, M.A.; Hardiman, O.; Chio, A.; Al-Chalabi, A.; Pasterkamp, R.J.; Veldink, J.H.; van den Berg, L.H. Amyotrophic lateral sclerosis. Lancet 2017, 390, 2084–2098. [Google Scholar] [CrossRef]
- Oskarsson, B.; Gendron, T.F.; Staff, N.P. Amyotrophic Lateral Sclerosis: An Update for 2018. Mayo Clin. Proc. 2018, 93, 1617–1628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hartzfeld, D.E.H.; Siddique, N.; Victorson, D.; O’Neill, S.; Kinsley, L.; Siddique, T. Reproductive decision-making among individuals at risk for familial amyotrophic lateral sclerosis. Amyotroph. Lateral Scler. Front. Degener. 2015, 16, 114–119. [Google Scholar] [CrossRef]
- Chia, R.; Chiò, A.; Traynor, B.J. Novel genes associated with amyotrophic lateral sclerosis: Diagnostic and clinical implications. Lancet Neurol. 2018, 17, 94–102. [Google Scholar] [CrossRef]
- Nguyen, H.P.; Van Broeckhoven, C.; van der Zee, J. ALS Genes in the Genomic Era and their Implications for FTD. Trends Genet. 2018, 34, 404–423. [Google Scholar] [CrossRef] [Green Version]
- Jaiswal, M.K. Riluzole and edaravone: A tale of two amyotrophic lateral sclerosis drugs. Med. Res. Rev. 2019, 39, 733–748. [Google Scholar] [CrossRef]
- Joilin, G.; Leigh, P.N.; Newbury, S.F.; Hafezparast, M. An Overview of MicroRNAs as Biomarkers of ALS. Front. Neurol. 2019, 10, 186. [Google Scholar] [CrossRef] [Green Version]
- Haramati, S.; Chapnik, E.; Sztainberg, Y.; Eilam, R.; Zwang, R.; Gershoni, N.; McGlinn, E.; Heiser, P.; Wills, A.-M.; Wirguin, I.; et al. miRNA malfunction causes spinal motor neuron disease. Proc. Natl. Acad. Sci. USA 2010, 107, 13111–13116. [Google Scholar] [CrossRef] [Green Version]
- De Paola, E.; Verdile, V.; Paronetto, M.P. Dysregulation of microRNA metabolism in motor neuron diseases: Novel biomarkers and potential therapeutics. Non Coding RNA Res. 2019, 4, 15–22. [Google Scholar] [CrossRef]
- Emde, A.; Eitan, C.; Liou, L.; Libby, R.T.; Rivkin, N.; Magen, I.; Reichenstein, I.; Oppenheim, H.; Eilam, R.; Silvestroni, A.; et al. Dysregulated mi RNA biogenesis downstream of cellular stress and ALS-causing mutations: A new mechanism for ALS. EMBO J. 2015, 34, 2633–2651. [Google Scholar] [CrossRef] [Green Version]
- Aravin, A.A.; Lagos-Quintana, M.; Yalcin, A.; Zavolan, M.; Marks, D.; Snyder, B.; Gaasterland, T.; Meyer, J.; Tuschl, T. The Small RNA Profile during Drosophila melanogaster Development. Dev. Cell 2003, 5, 337–350. [Google Scholar] [CrossRef] [Green Version]
- Amores, A.; Force, A.; Yan, Y.-L.; Joly, L.; Amemiya, C.; Fritz, A.; Ho, R.K.; Langeland, J.; Prince, V.; Wang, Y.-L.; et al. Zebrafish hox Clusters and Vertebrate Genome Evolution. Science 1998, 282, 1711–1714. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, X. miRDB: An online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020, 48, D127–D131. [Google Scholar] [CrossRef] [Green Version]
- Paul, S.; Vázquez, L.A.B.; Uribe, S.P.; Reyes-Pérez, P.R.; Sharma, A. Current Status of microRNA-Based Therapeutic Approaches in Neurodegenerative Disorders. Cells 2020, 9, 1698. [Google Scholar] [CrossRef]
- Filipowicz, W.; Bhattacharyya, S.N.; Sonenberg, N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat. Rev. Genet. 2008, 9, 102–114. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [Green Version]
- Fu, G.; Brkić, J.; Hayder, H.; Peng, C. MicroRNAs in Human Placental Development and Pregnancy Complications. Int. J. Mol. Sci. 2013, 14, 5519–5544. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.; Han, J.; Yeom, K.-H.; Jin, H.; Kim, V. Drosha in Primary MicroRNA Processing. In Cold Spring Harbor Symposia on Quantitative Biology; Cold Spring Harbor Laboratory Press: Long Island, NY, USA, 2006; Volume 71, pp. 51–57. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.; Jeon, K.; Lee, J.; Kim, S.; Kim, V.N. MicroRNA maturation: Stepwise processing and subcellular localization. EMBO J. 2002, 21, 4663–4670. [Google Scholar] [CrossRef] [Green Version]
- Huang, W. MicroRNAs: Biomarkers, Diagnostics, and Therapeutics. Bioinform. MicroRNA Res. 2017, 1617, 57–67. [Google Scholar] [CrossRef]
- Kim, Y.K.; Kim, V.N. Processing of intronic microRNAs. EMBO J. 2007, 26, 775–783. [Google Scholar] [CrossRef] [Green Version]
- Hayes, J.; Peruzzi, P.P.; Lawler, S. MicroRNAs in cancer: Biomarkers, functions and therapy. Trends Mol. Med. 2014, 20, 460–469. [Google Scholar] [CrossRef]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef]
- Zeng, Y.; Yi, R.; Cullen, B.R. MicroRNAs and small interfering RNAs can inhibit mRNA expression by similar mechanisms. Proc. Natl. Acad. Sci. USA 2003, 100, 9779–9784. [Google Scholar] [CrossRef] [Green Version]
- Bohnsack, M.T.; Czaplinski, K.; Görlich, D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA 2004, 10, 185–191. [Google Scholar] [CrossRef] [Green Version]
- Yi, R.; Qin, Y.; Macara, I.G.; Cullen, B.R. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003, 17, 3011–3016. [Google Scholar] [CrossRef] [Green Version]
- Lund, E.; Güttinger, S.; Calado, A.; Dahlberg, J.E.; Kutay, U. Nuclear Export of MicroRNA Precursors. Science 2004, 303, 95–98. [Google Scholar] [CrossRef] [Green Version]
- Tanzer, A.; Stadler, P.F. Molecular Evolution of a MicroRNA Cluster. J. Mol. Biol. 2004, 339, 327–335. [Google Scholar] [CrossRef]
- Grishok, A.; Pasquinelli, A.E.; Conte, D.; Li, N.; Parrish, S.; Ha, I.; Baillie, D.L.; Fire, A.; Ruvkun, G.; Mello, C.C. Genes and Mechanisms Related to RNA Interference Regulate Expression of the Small Temporal RNAs that Control C. elegans Developmental Timing. Cell 2001, 106, 23–34. [Google Scholar] [CrossRef] [Green Version]
- Hutvagner, G.; Zamore, P.D. A microRNA in a Multiple-Turnover RNAi Enzyme Complex. Science 2002, 297, 2056–2060. [Google Scholar] [CrossRef] [Green Version]
- Utsuyama, M.; Hirokawa, K.; Mancini, C.; Brunelli, R.; Leter, G.; Doria, G. Differential effects of gonadectomy on thymic stromal cells in promoting T cell differentiation in mice. Mech. Ageing Dev. 1995, 81, 107–117. [Google Scholar] [CrossRef]
- Hammond, S.M.; Bernstein, E.; Beach, D.; Hannon, G.J. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 2000, 404, 293–296. [Google Scholar] [CrossRef] [PubMed]
- Martinez, J.; Patkaniowska, A.; Urlaub, H.; Lührmann, R.; Tuschl, T. Single-Stranded Antisense siRNAs Guide Target RNA Cleavage in RNAi. Cell 2002, 110, 563–574. [Google Scholar] [CrossRef] [Green Version]
- Schwarz, D.S.; Hutvagner, G.; Haley, B.; Zamore, P.D. Evidence that siRNAs Function as Guides, Not Primers, in the Drosophila and Human RNAi Pathways. Mol. Cell 2002, 10, 537–548. [Google Scholar] [CrossRef]
- Lagos-Quintana, M.; Rauhut, R.; Meyer, J.; Borkhardt, A.; Tuschl, T. New microRNAs from mouse and human. RNA 2003, 9, 175–179. [Google Scholar] [CrossRef] [Green Version]
- De Rie, D.; Abugessaisa, I.; Alam, T.; Arner, E.; Arner, P.; Ashoor, H.; Åström, G.; Babina, M.; Bertin, N.; Burroughs, A.M.; et al. An integrated expression atlas of miRNAs and their promoters in human and mouse. Nat. Biotechnol. 2017, 35, 872–878. [Google Scholar] [CrossRef]
- Kabekkodu, S.; Shukla, V.; Varghese, V.K.; Souza, J.D.; Chakrabarty, S.; Satyamoorthy, K. Clustered miRNAs and their role in biological functions and diseases. Biol. Rev. 2018, 93, 1955–1986. [Google Scholar] [CrossRef]
- Makarova, J.A.; Shkurnikov, M.U.; Wicklein, D.; Lange, T.; Samatov, T.R.; Turchinovich, A.A.; Tonevitsky, A.G. Intracellular and extracellular microRNA: An update on localization and biological role. Prog. Histochem. Cytochem. 2016, 51, 33–49. [Google Scholar] [CrossRef]
- Benigni, M.; Ricci, C.; Jones, A.R.; Giannini, F.; Al-Chalabi, A.; Battistini, S. Identification of miRNAs as Potential Biomarkers in Cerebrospinal Fluid from Amyotrophic Lateral Sclerosis Patients. NeuroMolecular Med. 2016, 18, 551–560. [Google Scholar] [CrossRef]
- Pegoraro, V.; Merico, A.; Angelini, C. Micro-RNAs in ALS muscle: Differences in gender, age at onset and disease duration. J. Neurol. Sci. 2017, 380, 58–63. [Google Scholar] [CrossRef] [Green Version]
- Pegoraro, V.; Merico, A.; Angelini, C. MyomiRNAs Dysregulation in ALS Rehabilitation. Brain Sci. 2019, 9, 8. [Google Scholar] [CrossRef] [Green Version]
- Freischmidt, A.; Müller, K.; Zondler, L.; Weydt, P.; Volk, A.E.; Božič, A.L.; Walter, M.; Bonin, M.; Mayer, B.; Von Arnim, C.A.F.; et al. Serum microRNAs in patients with genetic amyotrophic lateral sclerosis and pre-manifest mutation carriers. Brain 2014, 137, 2938–2950. [Google Scholar] [CrossRef] [Green Version]
- Parisi, C.; Arisi, I.; D’Ambrosi, N.; Storti, A.E.; Brandi, R.; D’Onofrio, M.; Volonté, C. Dysregulated microRNAs in amyotrophic lateral sclerosis microglia modulate genes linked to neuroinflammation. Cell Death Dis. 2013, 4, e959. [Google Scholar] [CrossRef]
- Scott, C.E.; Wynn, S.L.; Sesay, A.; Cruz, C.; Cheung, M.; Gomez-Gaviro, M.V.; Booth, S.; Gao, B.; Cheah, K.S.E.; Lovell-Badge, R.; et al. SOX9 induces and maintains neural stem cells. Nat. Neurosci. 2010, 13, 1181–1189. [Google Scholar] [CrossRef]
- Hoye, M.L.; Regan, M.R.; Jensen, L.A.; Lake, A.M.; Reddy, L.V.; Vidensky, S.; Richard, J.-P.; Maragakis, N.J.; Rothstein, J.D.; Dougherty, J.D.; et al. Motor neuron-derived microRNAs cause astrocyte dysfunction in amyotrophic lateral sclerosis. Brain 2018, 141, 2561–2575. [Google Scholar] [CrossRef]
- Li, C.; Chen, Y.; Chen, X.; Wei, Q.; Cao, B.; Shang, H. Downregulation of MicroRNA-193b-3p Promotes Autophagy and Cell Survival by Targeting TSC1/mTOR Signaling in NSC-34 Cells. Front. Mol. Neurosci. 2017, 10, 160. [Google Scholar] [CrossRef] [Green Version]
- Rohm, M.; May, C.; Marcus, K.; Steinbach, S.; Theis, V.; Theiss, C.; Matschke, V. The microRNA miR-375-3p and the Tumor Suppressor NDRG2 are Involved in Sporadic Amyotrophic Lateral Sclerosis. Cell. Physiol. Biochem. 2019, 52, 1412–1426. [Google Scholar] [CrossRef]
- Campos-Melo, D.; Droppelmann, C.A.; He, Z.; Volkening, K.; Strong, M.J. Altered microRNA expression profile in amyotrophic lateral sclerosis: A role in the regulation of NFL mRNA levels. Mol. Brain 2013, 6, 26. [Google Scholar] [CrossRef] [Green Version]
- De Santis, R.; Santini, L.; Colantoni, A.; Peruzzi, G.; de Turris, V.; Alfano, V.; Bozzoni, I.; Rosa, A. FUS Mutant Human Motoneurons Display Altered Transcriptome and microRNA Pathways with Implications for ALS Pathogenesis. Stem Cell Rep. 2017, 9, 1450–1462. [Google Scholar] [CrossRef] [Green Version]
- Varcianna, A.; Myszczynska, M.A.; Castelli, L.M.; O’Neill, B.; Kim, Y.; Talbot, J.; Nyberg, S.; Nyamali, I.; Heath, P.R.; Stopford, M.J.; et al. Micro-RNAs secreted through astrocyte-derived extracellular vesicles cause neuronal network degeneration in C9orf72 ALS. EBioMedicine 2019, 40, 626–635. [Google Scholar] [CrossRef] [Green Version]
- De Felice, B.; Manfellotto, F.; Fiorentino, G.; Annunziata, A.; Biffali, E.; Pannone, R.; Federico, A. Wide-Ranging Analysis of MicroRNA Profiles in Sporadic Amyotrophic Lateral Sclerosis Using Next-Generation Sequencing. Front. Genet. 2018, 9, 310. [Google Scholar] [CrossRef]
- Raheja, R.; Regev, K.; Healy, B.C.; Mazzola, M.A.; Beynon, V.; Von Glehn, F.; Paul, A.; Diaz-Cruz, C.; Gholipour, T.; Glanz, B.I.; et al. Correlating serum micrornas and clinical parameters in amyotrophic lateral sclerosis. Muscle Nerve 2018, 58, 261–269. [Google Scholar] [CrossRef]
- Helferich, A.M.; Brockmann, S.J.; Reinders, J.; Deshpande, D.; Holzmann, K.; Brenner, D.; Andersen, P.M.; Petri, S.; Thal, D.; Michaelis, J.; et al. Dysregulation of a novel miR-1825/TBCB/TUBA4A pathway in sporadic and familial ALS. Cell. Mol. Life Sci. 2018, 75, 4301–4319. [Google Scholar] [CrossRef]
- de Andrade, H.M.; de Albuquerque, M.; Avansini, S.; Rocha, C.D.S.; Dogini, D.; Nucci, A.; Carvalho, B.; Lopes-Cendes, I.; França, M.C. MicroRNAs-424 and 206 are potential prognostic markers in spinal onset amyotrophic lateral sclerosis. J. Neurol. Sci. 2016, 368, 19–24. [Google Scholar] [CrossRef]
- De Felice, B.; Annunziata, A.; Fiorentino, G.; Borra, M.; Biffali, E.; Coppola, C.; Cotrufo, R.; Brettschneider, J.; Giordana, M.L.; Dalmay, T.; et al. miR-338-3p is over-expressed in blood, CFS, serum and spinal cord from sporadic amyotrophic lateral sclerosis patients. Neurogenetics 2014, 15, 243–253. [Google Scholar] [CrossRef]
- Waller, R.; Goodall, E.; Milo, M.; Cooper-Knock, J.; Da Costa, M.; Hobson, E.; Kazoka, M.; Wollff, H.; Heath, P.R.; Shaw, P.; et al. Serum miRNAs miR-206, 143–3p and 374b-5p as potential biomarkers for amyotrophic lateral sclerosis (ALS). Neurobiol. Aging 2017, 55, 123–131. [Google Scholar] [CrossRef]
- Nolan, K.; Mitchem, M.R.; Jimenez-Mateos, E.M.; Henshall, D.C.; Concannon, C.G.; Prehn, J.H.M. Increased Expression of MicroRNA-29a in ALS Mice: Functional Analysis of Its Inhibition. J. Mol. Neurosci. 2014, 53, 231–241. [Google Scholar] [CrossRef]
- Russell, A.P.; Wada, S.; Vergani, L.; Hock, M.B.; Lamon, S.; Léger, B.; Ushida, T.; Cartoni, R.; Wadley, G.D.; Hespel, P.; et al. Disruption of skeletal muscle mitochondrial network genes and miRNAs in amyotrophic lateral sclerosis. Neurobiol. Dis. 2013, 49, 107–117. [Google Scholar] [CrossRef]
- Zhou, F.; Guan, Y.; Chen, Y.; Zhang, C.; Yu, L.; Gao, H.; Du, H.; Liu, B.; Wang, X. miRNA-9 expression is upregulated in the spinal cord of G93A-SOD1 transgenic mice. Int. J. Clin. Exp. Pathol. 2013, 6, 1826–1838. [Google Scholar]
- Parisi, C.; Napoli, G.; Pelegrin, P.; Volonté, C. M1 and M2 Functional Imprinting of Primary Microglia: Role of P2X7 Activation and miR-125b. Mediat. Inflamm. 2016, 2016, 2989548. [Google Scholar] [CrossRef] [Green Version]
- Zhou, F.; Zhang, C.; Guan, Y.; Chen, Y.; Lu, Q.; Jie, L.; Gao, H.; Du, H.; Zhang, H.; Liu, Y.; et al. Screening the expression characteristics of several miRNAs in G93A-SOD1 transgenic mouse: Altered expression of miRNA-124 is associated with astrocyte differentiation by targeting Sox2 and Sox9. J. Neurochem. 2017, 145, 51–67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, C.-C.; Uchiyama, Y.; Hiyama, A.; Gajghate, S.; Shapiro, I.M.; Risbud, M.V. PI3K/AKT regulates aggrecan gene expression by modulating Sox9 expression and activity in nucleus pulposus cells of the intervertebral disc. J. Cell. Physiol. 2009, 221, 668–676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosenblum, L.T.; Trotti, D. EAAT2 and the Molecular Signature of Amyotrophic Lateral Sclerosis. Glial Amino Acid Transp. 2017, 16, 117–136. [Google Scholar] [CrossRef]
- Malik, A.R.; Willnow, T.E. Excitatory Amino Acid Transporters in Physiology and Disorders of the Central Nervous System. Int. J. Mol. Sci. 2019, 20, 5671. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.; Kundu, M.; Viollet, B.; Guan, K.-L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 2011, 13, 132–141. [Google Scholar] [CrossRef] [Green Version]
- Barmada, S.J.; Serio, A.; Arjun, A.; Bilican, B.; Daub, A.; Ando, D.; Tsvetkov, A.S.; Pleiss, M.; Li, X.; Peisach, D.; et al. Autophagy induction enhances TDP43 turnover and survival in neuronal ALS models. Nat. Chem. Biol. 2014, 10, 677–685. [Google Scholar] [CrossRef] [Green Version]
- Williams, A.H.; Valdez, G.; Moresi, V.; Qi, X.; McAnally, J.; Elliott, J.L.; Bassel-Duby, R.; Sanes, J.R.; Olson, E.N. MicroRNA-206 Delays ALS Progression and Promotes Regeneration of Neuromuscular Synapses in Mice. Science 2009, 326, 1549–1554. [Google Scholar] [CrossRef] [Green Version]
- Toivonen, J.M.; Manzano, R.; Oliván, S.; Zaragoza, P.; García-Redondo, A.; Osta, R. MicroRNA-206: A Potential Circulating Biomarker Candidate for Amyotrophic Lateral Sclerosis. PLoS ONE 2014, 9, e89065. [Google Scholar] [CrossRef] [Green Version]
- De Felice, B.; Guida, M.; Guida, M.; Coppola, C.; De Mieri, G.; Cotrufo, R. A miRNA signature in leukocytes from sporadic amyotrophic lateral sclerosis. Gene 2012, 508, 35–40. [Google Scholar] [CrossRef]
- Chen, Y.; Wei, Q.; Chen, X.; Li, C.; Cao, B.; Ou, R.; Hadano, S.; Shang, H.-F. Aberration of miRNAs Expression in Leukocytes from Sporadic Amyotrophic Lateral Sclerosis. Front. Mol. Neurosci. 2016, 9, 69. [Google Scholar] [CrossRef] [Green Version]
- Drory, V.E.; Goltsman, E.; Reznik, J.; Mosek, A.; Korczyn, A. The value of muscle exercise in patients with amyotrophic lateral sclerosis. J. Neurol. Sci. 2001, 191, 133–137. [Google Scholar] [CrossRef]
- Alexander, M.S.; Kunkel, L.M. Skeletal Muscle MicroRNAs: Their Diagnostic and Therapeutic Potential in Human Muscle Diseases. J. Neuromuscul. Dis. 2015, 2, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Horak, M.; Novak, J.; Bienertova-Vasku, J. Muscle-specific microRNAs in skeletal muscle development. Dev. Biol. 2016, 410, 1–13. [Google Scholar] [CrossRef]
- Rothstein, J.D. Therapeutic horizons for amyotrophic lateral sclerosis. Curr. Opin. Neurobiol. 1996, 6, 679–687. [Google Scholar] [CrossRef]
- Botta-Orfila, T.; Morató, X.; Compta, Y.; Lozano, J.J.; Falgàs, N.; Valldeoriola, F.; Pont-Sunyer, C.; Vilas, D.; Mengual, L.; Fernández, M.; et al. Identification of blood serum micro-RNAs associated with idiopathic andLRRK2Parkinson’s disease. J. Neurosci. Res. 2014, 92, 1071–1077. [Google Scholar] [CrossRef]
- Daniels, S.I.; Sille, F.; Goldbaum, A.; Yee, B.; Key, E.F.; Zhang, L.; Smith, M.T.; Thomas, R. Improving Power to Detect Changes in Blood miRNA Expression by Accounting for Sources of Variability in Experimental Designs. Cancer Epidemiol. Prev. Biomark. 2014, 23, 2658–2666. [Google Scholar] [CrossRef] [Green Version]
- Ma, Z.; Sun, X.; Xu, D.; Xiong, Y.; Zuo, B. MicroRNA, miR-374b, directly targets Myf6 and negatively regulates C2C12 myoblasts differentiation. Biochem. Biophys. Res. Commun. 2015, 467, 670–675. [Google Scholar] [CrossRef]
- Conti, A.; Aguennouz, M.; La Torre, D.; Tomasello, C.; Cardali, S.M.; Angileri, F.; Maio, F.; Cama, A.; Germanò, A.; Vita, G.; et al. miR-21 and 221 upregulation and miR-181b downregulation in human grade II–IV astrocytic tumors. J. Neuro Oncol. 2009, 93, 325–332. [Google Scholar] [CrossRef]
- Wijesekera Lokesh, C.; Nigel Leigh, P. Amyotrophic lateral sclerosis. Orphanet J. Rare Dis. 2009, 4, 1–22. [Google Scholar] [CrossRef] [Green Version]
- Imbalzano, A.N.; Jones, S.N. Loss of miRNA biogenesis induces p19Arf-p53 signaling and senescence in primary cells. J. Cell Biol. 2008, 181, 1055–1063. [Google Scholar] [CrossRef] [Green Version]
- Kuehbacher, A.; Urbich, C.; Zeiher, A.M.; Dimmeler, S. Role of Dicer and Drosha for Endothelial MicroRNA Expression and Angiogenesis. Circ. Res. 2007, 101, 59–68. [Google Scholar] [CrossRef] [Green Version]
- Forterre, A.; Komuro, H.; Aminova, S.; Harada, M. A Comprehensive Review of Cancer MicroRNA Therapeutic Delivery Strategies. Cancers 2020, 12, 1852. [Google Scholar] [CrossRef]
- Wang, Z. The Concept of Multiple-Target Anti-miRNA Antisense Oligonucleotide Technology. MicroRNA Cancer 2010, 676, 51–57. [Google Scholar] [CrossRef]
- Veedu, R.N.; Wengel, J. Locked Nucleic Acids: Promising Nucleic Acid Analogs for Therapeutic Applications. Chem. Biodivers. 2010, 7, 536–542. [Google Scholar] [CrossRef]
- Yue, J. miRNA and vascular cell movement. Adv. Drug Deliv. Rev. 2011, 63, 616–622. [Google Scholar] [CrossRef] [Green Version]
- Wen, D.; Danquah, M.; Chaudhary, A.K.; Mahato, R.I. Small molecules targeting microRNA for cancer therapy: Promises and obstacles. J. Control. Release 2015, 219, 237–247. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z. The Guideline of the Design and Validation of MiRNA Mimics. MicroRNA Cancer 2011, 676, 211–223. [Google Scholar] [CrossRef]
- Sureban, S.M.; May, R.; Mondalek, F.G.; Qu, D.; Ponnurangam, S.; Pantazis, P.; Anant, S.; Ramanujam, R.P.; Houchen, C.W. Nanoparticle-based delivery of siDCAMKL-1 increases microRNA-144 and inhibits colorectal cancer tumor growth via a Notch-1 dependent mechanism. J. Nanobiotechnol. 2011, 9, 40. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Yu, B.; Ren, W.; Mo, X.; Zhou, C.; He, H.; Jia, H.; Wang, L.; Jacob, S.T.; Lee, R.J.; et al. Enhanced hepatic delivery of siRNA and microRNA using oleic acid based lipid nanoparticle formulations. J. Control Release 2013, 172, 690–698. [Google Scholar] [CrossRef]
- Davis, S.; Lollo, B.; Freier, S.; Esau, C. Improved targeting of miRNA with antisense oligonucleotides. Nucleic Acids Res. 2006, 34, 2294–2304. [Google Scholar] [CrossRef]
- Elmén, J.; Lindow, M.; Silahtaroglu, A.; Bak, M.; Christensen, M.; Lind-Thomsen, A.; Hedtjärn, M.; Hansen, J.B.; Hansen, H.F.; Straarup, E.M.; et al. Antagonism of microRNA-122 in mice by systemically administered LNA-antimiR leads to up-regulation of a large set of predicted target mRNAs in the liver. Nucleic Acids Res. 2008, 36, 1153–1162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stenvang, J.; Petri, A.; Lindow, M.; Obad, S.; Kauppinen, S. Inhibition of microRNA function by antimiR oligonucleotides. Silence 2012, 3, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Gao, D.-Y.; Huang, L. In vivo delivery of miRNAs for cancer therapy: Challenges and strategies. Adv. Drug Deliv. Rev. 2015, 81, 128–141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simonson, B. MicroRNA Therapeutics: The Next Magic Bullet? Mini Rev. Med. Chem. 2015, 15, 467–474. [Google Scholar] [CrossRef]
- Pereira, D.; Rodrigues, P.; Borralho, P.; Rodrigues, C. Delivering the promise of miRNA cancer therapeutics. Drug Discov. Today 2013, 18, 282–289. [Google Scholar] [CrossRef]
- Sun, X.; Guo, Q.; Wei, W.; Robertson, S.; Yuan, Y.; Luo, X. Current Progress on MicroRNA-Based Gene Delivery in the Treatment of Osteoporosis and Osteoporotic Fracture. Int. J. Endocrinol. 2019, 2019, 6782653. [Google Scholar] [CrossRef] [Green Version]
- Yang, N. An overview of viral and nonviral delivery systems for microRNA. Int. J. Pharm. Investig. 2015, 5, 179–181. [Google Scholar] [CrossRef]
- Myoung, S.; Kasinski, A.L. Strategies for Safe and Targeted Delivery of MicroRNA Therapeutics. MicroRNAs Dis. Disord. 2019, 386–415. [Google Scholar] [CrossRef] [Green Version]
- Ojala, D.S.; Amara, D.P.; Schaffer, D.V. Adeno-Associated Virus Vectors and Neurological Gene Therapy. Neuroscientist 2015, 21, 84–98. [Google Scholar] [CrossRef]
- Martier, R.; Liefhebber, J.M.; Miniarikova, J.; van der Zon, T.; Snapper, J.; Kolder, I.; Petry, H.; van Deventer, S.J.; Evers, M.M.; Konstantinova, P. Artificial MicroRNAs Targeting C9orf72 Can Reduce Accumulation of Intra-nuclear Transcripts in ALS and FTD Patients. Mol. Ther. Nucleic Acids 2019, 14, 593–608. [Google Scholar] [CrossRef] [Green Version]
- Borel, F.; Gernoux, G.; Sun, H.; Stock, R.; Blackwood, M.; Brown, R.H.; Mueller, C. Safe and effective superoxide dismutase 1 silencing using artificial microRNA in macaques. Sci. Transl. Med. 2018, 10, eaau6414. [Google Scholar] [CrossRef] [Green Version]
- Bai, Z.; Wei, J.; Yu, C.; Han, X.; Qin, X.; Zhang, C.; Liao, W.; Li, L.; Huang, W. Non-viral nanocarriers for intracellular delivery of microRNA therapeutics. J. Mater. Chem. B 2019, 7, 1209–1225. [Google Scholar] [CrossRef]
- Wang, H.; Liu, S.; Jia, L.; Chu, F.; Zhou, Y.; He, Z.; Guo, M.; Chen, C.; Xu, L. Nanostructured lipid carriers for MicroRNA delivery in tumor gene therapy. Cancer Cell Int. 2018, 18, 101. [Google Scholar] [CrossRef]
- Bakhshandeh, B.; Soleimani, M.; Hafizi, M.; Ghaemi, N. A comparative study on nonviral genetic modifications in cord blood and bone marrow mesenchymal stem cells. Cytotechnology 2012, 64, 523–540. [Google Scholar] [CrossRef] [Green Version]
- Vaughan, H.J.; Green, J.J.; Tzeng, S.Y. Cancer-Targeting Nanoparticles for Combinatorial Nucleic Acid Delivery. Adv. Mater. 2020, 32, 1901081. [Google Scholar] [CrossRef]
- Blanco, E.; Shen, H.; Ferrari, M. Principles of nanoparticle design for overcoming biological barriers to drug delivery. Nat. Biotechnol. 2015, 33, 941–951. [Google Scholar] [CrossRef]
- Pecot, C.V.; Calin, G.A.; Coleman, R.L.; Lopez-Berestein, G.; Sood, A.K. RNA interference in the clinic: Challenges and future directions. Nat. Rev. Cancer 2011, 11, 59–67. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.W.L.; Paoletti, C.; Campisi, M.; Osaki, T.; Adriani, G.; Kamm, R.D.; Mattu, C.; Chiono, V. MicroRNA delivery through nanoparticles. J. Control. Release 2019, 313, 80–95. [Google Scholar] [CrossRef]
- Carter, M.; Shieh, J.C. Guide to Research Techniques in Neuroscience; Academic Press: Cambridge, MA, USA, 2015. [Google Scholar]
- Wen, M.M. Getting miRNA Therapeutics into the Target Cells for Neurodegenerative Diseases: A Mini-Review. Front. Mol. Neurosci. 2016, 9, 129. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Jiang, Y.; Peng, H.; Chen, Y.; Zhu, P.; Huang, Y. Recent progress in microRNA delivery for cancer therapy by non-viral synthetic vectors. Adv. Drug Deliv. Rev. 2015, 81, 142–160. [Google Scholar] [CrossRef]
- Magen, I.; Hornstein, E. Oligonucleotide-based therapy for neurodegenerative diseases. Brain Res. 2014, 1584, 116–128. [Google Scholar] [CrossRef] [PubMed]
- McDermott, A.M.; Heneghan, H.M.; Miller, N.; Kerin, M.J. The Therapeutic Potential of MicroRNAs: Disease Modulators and Drug Targets. Pharm. Res. 2011, 28, 3016–3029. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, K.; Breyne, K.; Ughetto, S.; Laurent, L.C.; Breakefield, X.O. RNA delivery by extracellular vesicles in mammalian cells and its applications. Nat. Rev. Mol. Cell Biol. 2020, 21, 585–606. [Google Scholar] [CrossRef] [PubMed]
- Guglielmi, L.; Nardella, M.; Musa, C.; Cifola, I.; Porru, M.; Cardinali, B.; Iannetti, I.; Di Pietro, C.; Bolasco, G.; Palmieri, V.; et al. Circulating miRNAs in Small Extracellular Vesicles Secreted by a Human Melanoma Xenograft in Mouse Brains. Cancers 2020, 12, 1635. [Google Scholar] [CrossRef]
- Munir, J.; Yoon, J.K.; Ryu, S. Therapeutic miRNA-Enriched Extracellular Vesicles: Current Approaches and Future Prospects. Cells 2020, 9, 2271. [Google Scholar] [CrossRef]
- Palmieri, V.; Lucchetti, D.; Gatto, I.; Maiorana, A.; Marcantoni, M.; Maulucci, G.; Papi, M.; Pola, R.; De Spirito, M.; Sgambato, A. Dynamic light scattering for the characterization and counting of extracellular vesicles: A powerful noninvasive tool. J. Nanoparticle Res. 2014, 16, 2583. [Google Scholar] [CrossRef]
- Moraes, F.C.; Pichon, C.; Letourneur, D.; Chaubet, F. miRNA Delivery by Nanosystems: State of the Art and Perspectives. Pharm. 2021, 13, 1901. [Google Scholar] [CrossRef]
- Novoselov, K.S.; Geim, A.K.; Morozov, S.V.; Jiang, D.; Zhang, Y.; Dubonos, S.V.; Grigorieva, I.V.; Firsov, A.A. Electric field effect in atomically thin carbon films. Science 2004, 306, 666–669. [Google Scholar] [CrossRef] [Green Version]
- Geim, A.K.; Novoselov, K.S. The rise of graphene. In Nanoscience and Technology: A Collection of Reviews from Nature Journals 11–19; World Scientific: Singapore, 2009; pp. 11–19. [Google Scholar] [CrossRef]
- Geim, A.K. Graphene: Status and Prospects. Science 2009, 324, 1530–1534. [Google Scholar] [CrossRef] [Green Version]
- Park, S.; Ruoff, R.S. Chemical methods for the production of graphenes. Nat. Nanotechnol. 2009, 4, 217–224. [Google Scholar] [CrossRef]
- Perini, G.; Palmieri, V.; Ciasca, G.; De Spirito, M.; Papi, M. Unravelling the Potential of Graphene Quantum Dots in Biomedicine and Neuroscience. Int. J. Mol. Sci. 2020, 21, 3712. [Google Scholar] [CrossRef]
- Asil, S.M.; Ahlawat, J.; Barroso, G.G.; Narayan, M. Nanomaterial based drug delivery systems for the treatment of neurodegenerative diseases. Biomater. Sci. 2020, 8, 4109–4128. [Google Scholar] [CrossRef]
- Safian, M.T.-U.; Haron, U.S.; Ibrahim, M.N.M. A review on bio-based graphene derived from biomass wastes. BioResources 2020, 15, 9756–9785. [Google Scholar] [CrossRef]
- Priyadarsini, S.; Mohanty, S.; Mukherjee, S.; Basu, S.; Mishra, M. Graphene and graphene oxide as nanomaterials for medicine and biology application. J. Nanostructure Chem. 2018, 8, 123–137. [Google Scholar] [CrossRef] [Green Version]
- Bitounis, D.; Ali-Boucetta, H.; Hong, B.H.; Min, D.-H.; Kostarelos, K. Prospects and Challenges of Graphene in Biomedical Applications. Adv. Mater. 2013, 25, 2258–2268. [Google Scholar] [CrossRef]
- Papi, M.; Palmieri, V.; Digiacomo, L.; Giulimondi, F.; Palchetti, S.; Ciasca, G.; Perini, G.; Caputo, D.; Cartillone, M.C.; Cascone, C.; et al. Converting the personalized biomolecular corona of graphene oxide nanoflakes into a high-throughput diagnostic test for early cancer detection. Nanoscale 2019, 11, 15339–15346. [Google Scholar] [CrossRef]
- Palmieri, V.; Barba, M.; Di Pietro, L.; Gentilini, S.; Braidotti, M.C.; Ciancico, C.; Bugli, F.; Ciasca, G.; Larciprete, R.; Lattanzi, W.; et al. Reduction and shaping of graphene-oxide by laser-printing for controlled bone tissue regeneration and bacterial killing. 2D Mater. 2017, 5, 015027. [Google Scholar] [CrossRef]
- Di Santo, R.; Quagliarini, E.; Palchetti, S.; Pozzi, D.; Palmieri, V.; Perini, G.; Papi, M.; Capriotti, A.L.; Laganà, A.; Caracciolo, G. Microfluidic-generated lipid-graphene oxide nanoparticles for gene delivery. Appl. Phys. Lett. 2019, 114, 233701. [Google Scholar] [CrossRef]
- Lu, C.H.; Yang, H.H.; Zhu, C.L.; Chen, X.; Chen, G.N. A Graphene Platform for Sensing Biomolecules. Angew. Chem. 2009, 121, 4879–4881. [Google Scholar] [CrossRef]
- Patil, A.J.; Vickery, J.L.; Scott, T.B.; Mann, S. Aqueous Stabilization and Self-Assembly of Graphene Sheets into Layered Bio-Nanocomposites using DNA. Adv. Mater. 2009, 21, 3159–3164. [Google Scholar] [CrossRef]
- Cui, L.; Chen, Z.; Zhu, Z.; Lin, X.; Chen, X.; Yang, C.J. Stabilization of ssRNA on Graphene Oxide Surface: An Effective Way to Design Highly Robust RNA Probes. Anal. Chem. 2013, 85, 2269–2275. [Google Scholar] [CrossRef]
- He, S.; Song, B.; Li, D.; Zhu, C.; Qi, W.; Wen, Y.; Wang, L.; Song, S.; Fang, H.; Fan, C. A Graphene Nanoprobe for Rapid, Sensitive, and Multicolor Fluorescent DNA Analysis. Adv. Funct. Mater. 2010, 20, 453–459. [Google Scholar] [CrossRef]
- Tang, L.; Chang, H.; Liu, Y.; Li, J. Duplex DNA/Graphene Oxide Biointerface: From Fundamental Understanding to Specific Enzymatic Effects. Adv. Funct. Mater. 2012, 22, 3083–3088. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, S.; Du, D.; Shao, Y.; Li, Z.; Wang, J.; Engelhard, M.H.; Li, J.; Lin, Y. Self assembly of acetylcholinesterase on a gold nanoparticles–graphene nanosheet hybrid for organophosphate pesticide detection using polyelectrolyte as a linker. J. Mater. Chem. 2011, 21, 5319–5325. [Google Scholar] [CrossRef]
- Lei, H.; Mi, L.; Zhou, X.; Chen, J.; Hu, J.; Guo, S.; Zhang, Y. Adsorption of double-stranded DNA to graphene oxide preventing enzymatic digestion. Nanoscale 2011, 3, 3888–3892. [Google Scholar] [CrossRef]
- Palmieri, V.; Di Pietro, L.; Perini, G.; Barba, M.; Parolini, O.; De Spirito, M.; Lattanzi, W.; Papi, M. Graphene Oxide Nano-Concentrators Selectively Modulate RNA Trapping According to Metal Cations in Solution. Front. Bioeng. Biotechnol. 2020, 8, 421. [Google Scholar] [CrossRef]
- Lu, C.-H.; Zhu, C.-L.; Li, J.; Liu, J.-J.; Chen, X.; Yang, H.-H. Using graphene to protect DNA from cleavage during cellular delivery. Chem. Commun. 2010, 46, 3116–3118. [Google Scholar] [CrossRef] [Green Version]
- Mohammadian, N.; Faridbod, F. ALS genosensing using DNA-hybridization electrochemical biosensor based on label-free immobilization of ssDNA on Sm2O3 NPs-rGO/PANI composite. Sensors Actuators B Chem. 2018, 275, 432–438. [Google Scholar] [CrossRef]
- Chua, C.K.; Pumera, M. Covalent chemistry on graphene. Chem. Soc. Rev. 2013, 42, 3222–3233. [Google Scholar] [CrossRef]
- Dreyer, D.R.; Park, S.; Bielawski, C.W.; Ruoff, R.S. The chemistry of graphene oxide. Chem. Soc. Rev. 2010, 39, 228–240. [Google Scholar] [CrossRef]
- Yan, L.; Zheng, Y.B.; Zhao, F.; Li, S.; Gao, X.; Xu, B.; Weiss, P.S.; Zhao, Y. Chemistry and physics of a single atomic layer: Strategies and challenges for functionalization of graphene and graphene-based materials. Chem. Soc. Rev. 2012, 41, 97–114. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Wan, J.; Zhang, S.; Zhang, Y.; Lee, S.-T.; Liu, Z. In Vivo Pharmacokinetics, Long-Term Biodistribution, and Toxicology of PEGylated Graphene in Mice. ACS Nano 2010, 5, 516–522. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Robinson, J.T.; Sun, X.; Dai, H. PEGylated Nanographene Oxide for Delivery of Water-Insoluble Cancer Drugs. J. Am. Chem. Soc. 2008, 130, 10876–10877. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, L.; Yang, X.; Shi, X.; Tan, X.; Peng, R.; Wang, J.; Liu, Z. Polyethylene Glycol and Polyethylenimine Dual-Functionalized Nano-Graphene Oxide for Photothermally Enhanced Gene Delivery. Small 2013, 9, 1989–1997. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Liu, M.; Zhang, L.; Huang, J.; Yao, J.; Zhang, Z. Polyethylenimine-functionalized graphene oxide as an efficient gene delivery vector. J. Mater. Chem. 2011, 21, 7736–7741. [Google Scholar] [CrossRef]
- Dong, H.; Ding, L.; Yan, F.; Ji, H.; Ju, H. The use of polyethylenimine-grafted graphene nanoribbon for cellular delivery of locked nucleic acid modified molecular beacon for recognition of microRNA. Biomaterials 2011, 32, 3875–3882. [Google Scholar] [CrossRef]
- Lungwitz, U.; Breunig, M.; Blunk, T.; Göpferich, A. Polyethylenimine-based non-viral gene delivery systems. Eur. J. Pharm. Biopharm. 2005, 60, 247–266. [Google Scholar] [CrossRef]
- Feng, L.; Zhang, S.; Liu, Z. Graphene based gene transfection. Nanoscale 2011, 3, 1252–1257. [Google Scholar] [CrossRef]
- Dong, H.; Dai, W.; Ju, H.; Lu, H.; Wang, S.; Xu, L.; Zhou, S.-F.; Zhang, Y.; Zhang, X. Multifunctional Poly(l-lactide)–Polyethylene Glycol-Grafted Graphene Quantum Dots for Intracellular MicroRNA Imaging and Combined Specific-Gene-Targeting Agents Delivery for Improved Therapeutics. ACS Appl. Mater. Interfaces 2015, 7, 11015–11023. [Google Scholar] [CrossRef]
- Tripathi, S.K.; Goyal, R.; Gupta, K.C.; Kumar, P. Functionalized graphene oxide mediated nucleic acid delivery. Carbon 2013, 51, 224–235. [Google Scholar] [CrossRef]
- Huang, Y.-P.; Hung, C.-M.; Hsu, Y.-C.; Zhong, C.-Y.; Wang, W.-R.; Chang, C.-C.; Lee, M.-J. Suppression of Breast Cancer Cell Migration by Small Interfering RNA Delivered by Polyethylenimine-Functionalized Graphene Oxide. Nanoscale Res. Lett. 2016, 11, 247. [Google Scholar] [CrossRef] [Green Version]
- Chlanda, A.; Kowiorski, K.; Małek, M.; Kijeńska-Gawrońska, E.; Bil, M.; Djas, M.; Strachowski, T.; Swieszkowski, W.; Lipińska, L. Morphology and Chemical Purity of Water Suspension of Graphene Oxide FLAKES Aged for 14 Months in Ambient Conditions. A Preliminary Study. Material 2021, 14, 4108. [Google Scholar] [CrossRef]
- Defteralı, Ç.; Verdejo, R.; Peponi, L.; Martín, E.D.; Martínez-Murillo, R.; López-Manchado, M.Á.; Vicario-Abejón, C. Thermally reduced graphene is a permissive material for neurons and astrocytes and de novo neurogenesis in the adult olfactory bulb in vivo. Biomaterials 2016, 82, 84–93. [Google Scholar] [CrossRef]
- Kumar, R.; Rauti, R.; Scaini, D.; Antman-Passig, M.; Meshulam, O.; Naveh, D.; Ballerini, L.; Shefi, O. Graphene-Based Nanomaterials for Neuroengineering: Recent Advances and Future Prospective. Adv. Funct. Mater. 2021, 31, 2104887. [Google Scholar] [CrossRef]
- Zhu, F.-D.; Hu, Y.-J.; Yu, L.; Zhou, X.-G.; Wu, J.-M.; Tang, Y.; Qin, D.-L.; Fan, Q.-Z.; Wu, A.-G. Nanoparticles: A Hope for the Treatment of Inflammation in CNS. Front. Pharmacol. 2021, 12, 1114. [Google Scholar] [CrossRef]
- Solanki, A.; Chueng, S.-T.; Yin, P.; Kappera, R.; Chhowalla, M.; Lee, K.-B. Axonal Alignment and Enhanced Neuronal Differentiation of Neural Stem Cells on Graphene-Nanoparticle Hybrid Structures. Adv. Mater. 2013, 25, 5477–5482. [Google Scholar] [CrossRef] [Green Version]
- Di Santo, R.; Digiacomo, L.; Palchetti, S.; Palmieri, V.; Perini, G.; Pozzi, D.; Papi, M.; Caracciolo, G. Microfluidic manufacturing of surface-functionalized graphene oxide nanoflakes for gene delivery. Nanoscale 2019, 11, 2733–2741. [Google Scholar] [CrossRef]
- Xiong, S.; Luo, J.; Wang, Q.; Li, Z.; Li, J.; Liu, Q.; Gao, L.; Fang, S.; Li, Y.; Pan, H.; et al. Targeted graphene oxide for drug delivery as a therapeutic nanoplatform against Parkinson’s disease. Biomater. Sci. 2021, 9, 1705–1715. [Google Scholar] [CrossRef]
- Pradhan, K.; Das, G.; Khan, J.; Gupta, V.; Barman, S.; Adak, A.; Ghosh, S. Neuro-Regenerative Choline-Functionalized Injectable Graphene Oxide Hydrogel Repairs Focal Brain Injury. ACS Chem. Neurosci. 2018, 10, 1535–1543. [Google Scholar] [CrossRef]


| MiRNA | Model | Function and Changes | Ref | |
|---|---|---|---|---|
| Animal model | miRNA-125b | SOD1-G93A mouse | Increase of TNFα transcription by interfering with the STAT3 pathway | [44] |
miRNA-124 | Aberrant modulation of astrocytic differentiation by targeting Sox2 and Sox9 | [45] | ||
miRNA-128 | Aberrant modulation of the excitatory amino acid transporter 2 | [46] | ||
miRNA-193b-3p | Stimulation of cell death by targeting tuberous sclerosis 1 (TSC1) | [47] | ||
miRNA-375-3p | Wobbler mouse | Inefficient inhibition of p53, increasing ROS production, and induction of cell death | [48] | |
| ALS patients | miRNA-146* | sALS | Interaction with NFL mRNA, and suppression of its expression | [49] |
miRNA-524-5p miRNA-582-3p  | sALS | Aberrant regulation of NFL mRNA 3′UTR | [49] | |
miRNA-375 | FUS mutant ALS | Aberrant targeting of the apoptotic factor ELAV-like protein 4 | [50] | |
miRNA-494-3p | C9orf72 mutant ALS | Aberrant regulation of semaphorin 3A | [51] | |
miRNA-223-3p miRNA-338-39  miRNA-326  | sALS | Altered homeostasis in the neuromuscular junction | [52] | |
miRNA-1 miRNA-19a-3p  miRNA-133a-3p  miRNA-133b  miRNA-144-5p  miRNA-192-3p  miRNA-192-5p  | fALS and sALS | Altered homeostasis in the serum | [53] | |
Let-7d3p miRNA-139-5p  miRNA-320a  miRNA-320c  miRNA-320b  miRNA-425-5p  | sALS and fALS | Altered homeostasis in the serum | [53] | |
miRNA-1825 | sALS and fALS | Overtranslation of tubulin-folding cofactor b, which contributes to the degradation of tubulin alpha-4A | [54] | |
miRNA-424 miRNA-206  | ALS | Altered homeostasis in the plasma and in the skeletal muscle | [55] | |
miRNA-338-3p | sALS | Altered homeostasis in leukocytes | [56] | |
miRNA-206 | sALS | Altered homeostasis in the serum (no significant change in the serum over time) | [57] | |
miRNA-143-3p miRNA-374b-5p  | sALS | Altered homeostasis in the serum (significant change in the serum over time) | [57] | |
miRNA-181a-5p miRNA-21-5p  miRNA-15b-5p miRNA-15b-5p | ALS | Alteration of apoptotic mechanism inhibition and cell proliferation | [40] | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Niccolini, B.; Palmieri, V.; De Spirito, M.; Papi, M. Opportunities Offered by Graphene Nanoparticles for MicroRNAs Delivery for Amyotrophic Lateral Sclerosis Treatment. Materials 2022, 15, 126. https://doi.org/10.3390/ma15010126
Niccolini B, Palmieri V, De Spirito M, Papi M. Opportunities Offered by Graphene Nanoparticles for MicroRNAs Delivery for Amyotrophic Lateral Sclerosis Treatment. Materials. 2022; 15(1):126. https://doi.org/10.3390/ma15010126
Chicago/Turabian StyleNiccolini, Benedetta, Valentina Palmieri, Marco De Spirito, and Massimiliano Papi. 2022. "Opportunities Offered by Graphene Nanoparticles for MicroRNAs Delivery for Amyotrophic Lateral Sclerosis Treatment" Materials 15, no. 1: 126. https://doi.org/10.3390/ma15010126







