Rosmarinic Acid-Loaded Polymeric Nanoparticles Prepared by Low-Energy Nano-Emulsion Templating: Formulation, Biophysical Characterization, and In Vitro Studies †
Abstract
:1. Introduction
2. Materials and Methods
2.1. Preparation of Polymeric Nano-Emulsions
2.2. Preparation of Polymeric Nanoparticles from Nano-Emulsion Templating
2.3. Physicochemical Characterization of Nanoparticles
2.4. Rosmarinic Acid (Ra) Quantification Assays
2.5. Encapsulation Efficiency (EE%)
2.6. Hyperspectral Microscopy and Dark-Field Imaging
2.7. Transmission Electron Microscopy (TEM)
2.8. In Vitro Drug Release Experiments
2.9. Mathematical Models
2.10. Protein Corona Formation
2.11. In Vitro Antioxidative Activity and EC50
2.12. In Vitro Cytotoxicity Test
2.13. Flow Cytometry
3. Results
3.1. Preparation of Polymeric Nanoparticles
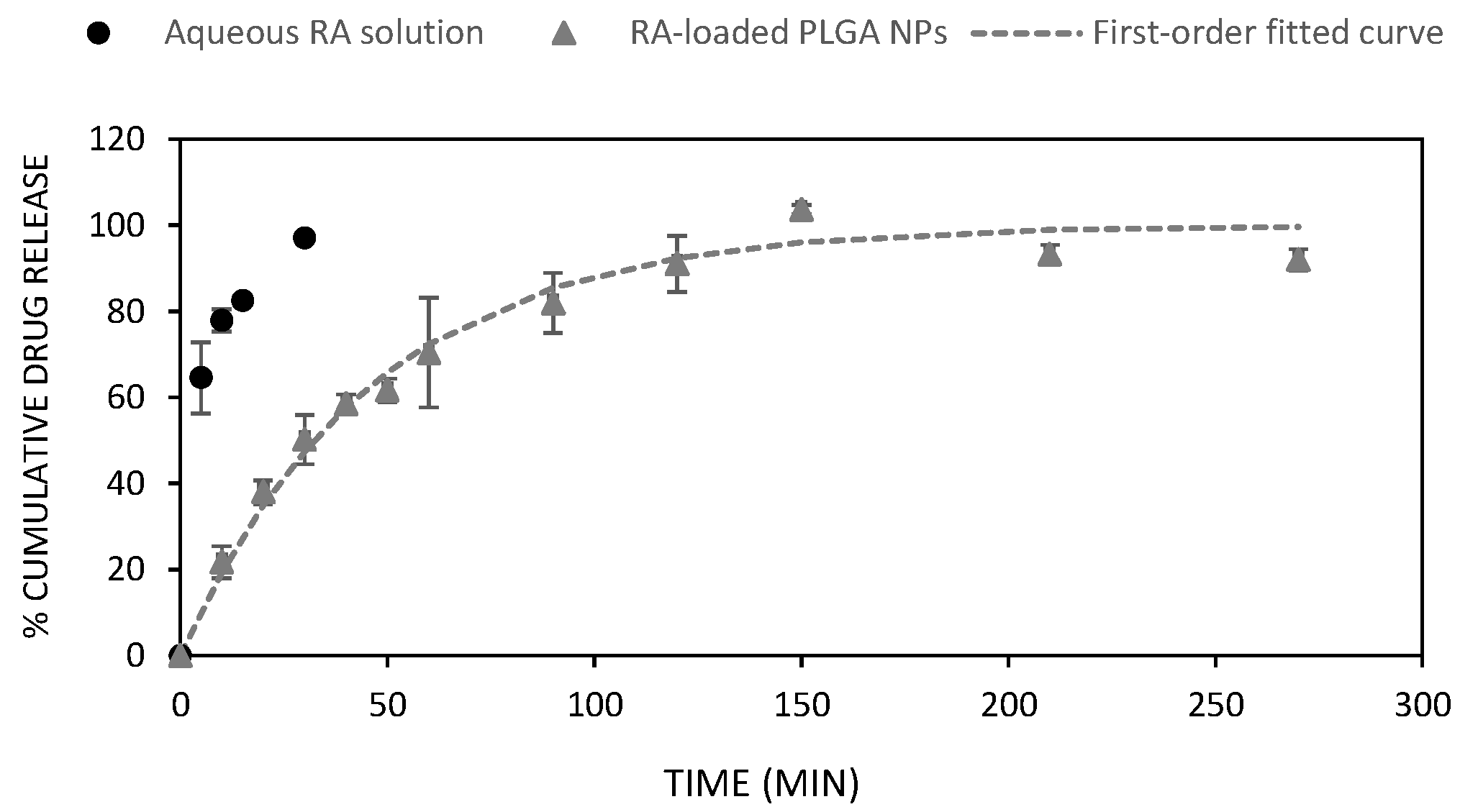
3.2. In Vitro Drug Release
3.3. Protein Corona (PC)
3.4. In Vitro Antioxidative Activity and EC50
3.5. Cytotoxicity
3.6. Cellular Uptake
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Harjo, B.; Wibowo, C.; Ng, K.M. Development of Natural Product Manufacturing Processes: Phytochemicals. Chem. Eng. Res. Des. 2004, 82, 1010–1028. [Google Scholar] [CrossRef]
- Panche, A.N.; Diwan, A.D.; Chandra, S.R. Flavonoids: An Overview. J. Nutr. Sci. 2016, 5, e47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holst, B.; Williamson, G. Nutrients and Phytochemicals: From Bioavailability to Bioefficacy beyond Antioxidants. Curr. Opin. Biotech. 2008, 19, 73–82. [Google Scholar] [CrossRef]
- Sulaiman, I.S.C.; Sukhi, S.; Mohamad, A. Roles of Phytochemicals in the Prevention and Treatment of Various Diseases. In Phytochemistry; Apple Academic Press: London, UK, 2018; pp. 147–164. ISBN 0429426194. [Google Scholar]
- Petersen, M. Rosmarinic Acid: New Aspects. Phytochem. Rev. 2013, 12, 207–227. [Google Scholar] [CrossRef]
- Petersen, M.; Simmonds, M.S.J. Rosmarinic Acid. Phytochemistry 2003, 62, 121–125. [Google Scholar] [CrossRef]
- Nagele, R.G.; D’Andrea, M.R.; Anderson, W.J.; Wang, H.-Y. Intracellular Accumulation of β-Amyloid1–42 in Neurons Is Facilitated by the A7 Nicotinic Acetylcholine Receptor in Alzheimer’s Disease. Neuroscience 2002, 110, 199–211. [Google Scholar] [CrossRef]
- Kumar, G.P.; Khanum, F. Neuroprotective Potential of Phytochemicals. Pharmacogn. Rev. 2012, 6, 81–90. [Google Scholar] [CrossRef] [Green Version]
- Hase, T.; Shishido, S.; Yamamoto, S.; Yamashita, R.; Nukima, H.; Taira, S.; Toyoda, T.; Abe, K.; Hamaguchi, T.; Ono, K.; et al. Rosmarinic Acid Suppresses Alzheimer’s Disease Development by Reducing Amyloid β Aggregation by Increasing Monoamine Secretion. Sci. Rep. 2019, 9, 8711. [Google Scholar] [CrossRef]
- Yamamoto, S.; Kayama, T.; Noguchi-Shinohara, M.; Hamaguchi, T.; Yamada, M.; Abe, K.; Kobayashi, S. Rosmarinic Acid Suppresses Tau Phosphorylation and Cognitive Decline by Downregulating the JNK Signaling Pathway. NPJ Sci. Food 2021, 5, 1. [Google Scholar] [CrossRef]
- Maher, P. The Potential of Flavonoids for the Treatment of Neurodegenerative Diseases. Int. J. Mol. Sci. 2019, 20, 3056. [Google Scholar] [CrossRef] [Green Version]
- De, R.; Jo, K.W.; Kim, K.-T. Influence of Molecular Structures on Fluorescence of Flavonoids and Their Detection in Mammalian Cells. Biomedicines 2022, 10, 1265. [Google Scholar] [CrossRef] [PubMed]
- Aqil, F.; Munagala, R.; Jeyabalan, J.; Vadhanam, M.v. Bioavailability of Phytochemicals and Its Enhancement by Drug Delivery Systems. Cancer Lett. 2013, 334, 133–141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rizwanullah, M.d.; Amin, S.; Mir, S.R.; Fakhri, K.U.; Rizvi Mohd, M.A. Phytochemical Based Nanomedicines against Cancer: Current Status and Future Prospects. J. Drug Target. 2017, 26, 731–752. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, R.; Srivastava, S.; Ghosh, S.; Khare, S.K. Phytochemical Delivery through Nanocarriers: A Review. Colloids Surf. B 2021, 197, 111389. [Google Scholar] [CrossRef]
- Li, Z.; Zhao, T.; Li, J.; Yu, Q.; Feng, Y.; Xie, Y.; Sun, P. Nanomedicine Based on Natural Products: Improving Clinical Application Potential. J. Nanomater. 2022, 2022, 3066613. [Google Scholar] [CrossRef]
- De, R.; Mahata, M.K.; Kim, K. Structure-Based Varieties of Polymeric Nanocarriers and Influences of Their Physicochemical Properties on Drug Delivery Profiles. Adv. Sci. 2022, 9, 2105373. [Google Scholar] [CrossRef]
- Begines, B.; Ortiz, T.; Pérez-Aranda, M.; Martínez, G.; Merinero, M.; Argüelles-Arias, F.; Alcudia, A. Polymeric Nanoparticles for Drug Delivery: Recent Developments and Future Prospects. Nanomaterials 2020, 10, 1403. [Google Scholar] [CrossRef]
- Giacalone, G.; Tsapis, N.; Mousnier, L.; Chacun, H.; Fattal, E. PLA-PEG Nanoparticles Improve the Anti-Inflammatory Effect of Rosiglitazone on Macrophages by Enhancing Drug Uptake Compared to Free Rosiglitazone. Materials 2018, 11, 1845. [Google Scholar] [CrossRef] [Green Version]
- Naskar, S.; Das, S.K.; Sharma, S.; Kuotsu, K. A Review on Designing Poly (Lactic-Co-Glycolic Acid) Nanoparticles as Drug Delivery Systems. Pharm. Nanotech. 2020, 9, 36–50. [Google Scholar] [CrossRef]
- Chatterjee, M.; Chanda, N. Formulation of PLGA Nano-Carrier: Specialized Modification for Cancer Therapeutic Applications. Mater. Adv. 2022, 3, 837–858. [Google Scholar] [CrossRef]
- Piotrowski-Daspit, A.S.; Kauffman, A.C.; Bracaglia, L.G.; Saltzman, W.M. Polymeric Vehicles for Nucleic Acid Delivery. Adv. Drug Deliv. Rev. 2020, 156, 119–132. [Google Scholar] [CrossRef] [PubMed]
- Zada, M.H.; Rottenberg, Y.; Domb, A.J. Peptide Loaded Polymeric Nanoparticles by Non-Aqueous Nanoprecipitation. J. Colloid Interface Sci. 2022, 622, 904–913. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.P.; Ramarao, P. Accumulated Polymer Degradation Products as Effector Molecules in Cytotoxicity of Polymeric Nanoparticles. Toxicol. Sci. 2013, 136, 131–143. [Google Scholar] [CrossRef]
- Håkansson, A.; Rayner, M. General Principles of Nanoemulsion Formation by High-Energy Mechanical Methods. In Nanoemulsions Formulation, Applications, and Characterization; Academic Press: Cambridge, MA, USA, 2018; pp. 103–139. ISBN 978-0-12-811838-2. [Google Scholar] [CrossRef]
- Solans, C.; Izquierdo, P.; Nolla, J.; Azemar, N.; Garcia-Celma, M.J. Nano-Emulsions. Curr. Opin. Colloid Interface Sci. 2005, 10, 102–110. [Google Scholar] [CrossRef]
- Solans, C.; Solé, I. Nano-Emulsions: Formation by Low-Energy Methods. Curr. Opin. Colloid Interface Sci. 2012, 17, 246–254. [Google Scholar] [CrossRef]
- Yang, Y.; Marshall-Breton, C.; Leser, M.E.; Sher, A.A.; McClements, D.J. Fabrication of Ultrafine Edible Emulsions: Comparison of High-Energy and Low-Energy Homogenization Methods. Food Hydrocoll. 2012, 29, 398–406. [Google Scholar] [CrossRef]
- Solè, I.; Maestro, A.; González, C.; Solans, C.; Gutiérrez, J.M. Optimization of Nano-Emulsion Preparation by Low-Energy Methods in an Ionic Surfactant System. Langmuir 2006, 22, 8326–8332. [Google Scholar] [CrossRef]
- Sadurní, N.; Solans, C.; Azemar, N.; García-Celma, M.J. Studies on the Formation of O/W Nano-Emulsions, by Low-Energy Emulsification Methods, Suitable for Pharmaceutical Applications. Eur. J. Pharm. Sci. 2005, 26, 438–445. [Google Scholar] [CrossRef]
- Klymchenko, A.S.; Liu, F.; Collot, M.; Anton, N. Dye-Loaded Nanoemulsions: Biomimetic Fluorescent Nanocarriers for Bioimaging and Nanomedicine. Adv. Health. Mater. 2021, 10, 2001289. [Google Scholar] [CrossRef]
- Sasikumar, A.; Kamalasanan, K. Nanomedicine for Prostate Cancer Using Nanoemulsion: A Review. J. Control. Release 2017, 260, 111–123. [Google Scholar] [CrossRef]
- Allen, T.M.; Cullis, P.R. Drug Delivery Systems: Entering the Mainstream. Science 2004, 303, 1818–1822. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fornaguera, C.; Grijalvo, S.; Galán, M.; Fuentes-Paniagua, E.; de la Mata, F.J.; Gómez, R.; Eritja, R.; Calderó, G.; Solans, C. Novel Non-Viral Gene Delivery Systems Composed of Carbosilane Dendron Functionalized Nanoparticles Prepared from Nano-Emulsions as Non-Viral Carriers for Antisense Oligonucleotides. Int. J. Pharm. 2015, 478, 113–123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fornaguera, C.; Dols-Perez, A.; Calderó, G.; García-Celma, M.J.; Camarasa, J.; Solans, C. PLGA Nanoparticles Prepared by Nano-Emulsion Templating Using Low-Energy Methods as Efficient Nanocarriers for Drug Delivery across the Blood–Brain Barrier. J. Control. Release 2015, 211, 134–143. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Negro, M.; González-Rubio, G.; Aicart, E.; Landfester, K.; Guerrero-Martínez, A.; Junquera, E. Insights into Colloidal Nanoparticle-Protein Corona Interactions for Nanomedicine Applications. Adv. Colloid Interface Sci. 2021, 289, 102366. [Google Scholar] [CrossRef]
- Li, H.; Wang, Y.; Tang, Q.; Yin, D.; Tang, C.; He, E.; Zou, L.; Peng, Q. The Protein Corona and Its Effects on Nanoparticle-Based Drug Delivery Systems. Acta Biomater. 2021, 129, 57–72. [Google Scholar] [CrossRef]
- Berrecoso, G.; Crecente-Campo, J.; Alonso, M.J. Unveiling the Pitfalls of the Protein Corona of Polymeric Drug Nanocarriers. Drug Deliv. Transl. Res. 2020, 10, 730–750. [Google Scholar] [CrossRef]
- Bertrand, N.; Grenier, P.; Mahmoudi, M.; Lima, E.M.; Appel, E.A.; Dormont, F.; Lim, J.-M.; Karnik, R.; Langer, R.; Farokhzad, O.C. Mechanistic Understanding of in Vivo Protein Corona Formation on Polymeric Nanoparticles and Impact on Pharmacokinetics. Nature Commun. 2017, 8, 777. [Google Scholar] [CrossRef]
- Corbo, C.; Molinaro, R.; Taraballi, F.; Toledano Furman, N.E.; Sherman, M.B.; Parodi, A.; Salvatore, F.; Tasciotti, E. Effects of the Protein Corona on Liposome-Liposome and Liposome-Cell Interactions. Int. J. Nanomed. 2016, 11, 3049–3063. [Google Scholar] [CrossRef] [Green Version]
- Monge, M.; Fornaguera, C.; Quero, C.; Dols-Perez, A.; Calderó, G.; Grijalvo, S.; García-Celma, M.J.; Rodríguez-Abreu, C.; Solans, C. Functionalized PLGA Nanoparticles Prepared by Nano-Emulsion Templating Interact Selectively with Proteins Involved in the Transport through the Blood-Brain Barrier. Eur. J. Pharm. Biopharm. 2020, 156, 155–164. [Google Scholar] [CrossRef]
- Yusuf, M.; Leung, K.; Morris, K.J.; Volpi, E.V. Comprehensive Cytogenomic Profile of the in Vitro Neuronal Model SH-SY5Y. Neurogenetics 2013, 14, 63–70. [Google Scholar] [CrossRef] [Green Version]
- Feng, J.; Rodríguez-Abreu, C.; Esquena, J.; Solans, C. A Concise Review on Nano-emulsion Formation by the Phase Inversion Composition (PIC) Method. J. Surfactants Deterg. 2020, 23, 677–685. [Google Scholar] [CrossRef]
- Higuchi, T. Mechanism of Sustained-Action Medication. Theoretical Analysis of Rate of Release of Solid Drugs Dispersed in Solid Matrices. J. Pharm. Sci. 1963, 52, 1145–1149. [Google Scholar] [CrossRef] [PubMed]
- Ritger, P.L.; Peppas, N.A. A Simple Equation for Description of Solute Release II. Fickian and Anomalous Release from Swellable Devices. J. Control. Release 1987, 5, 37–42. [Google Scholar] [CrossRef]
- Zhang, Y.; Huo, M.; Zhou, J.; Zou, A.; Li, W.; Yao, C.; Xie, S. DDSolver: An Add-in Program for Modeling and Comparison of Drug Dissolution Profiles. AAPS J 2010, 12, 263–271. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Bao, Y.; Ahire, J.; Chao, Y. Co-Encapsulation of Biodegradable Nanoparticles with Silicon Quantum Dots and Quercetin for Monitored Delivery. Adv. Health Mater. 2013, 2, 459–466. [Google Scholar] [CrossRef] [PubMed]
- Shubhra, Q.T.H.; Tóth, J.; Gyenis, J.; Feczkó, T. Surface Modification of HSA Containing Magnetic PLGA Nanoparticles by Poloxamer to Decrease Plasma Protein Adsorption. Colloids Surf. B 2014, 122, 529–536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, Y.; Chen, Y.; Yu, T.; Guo, Y.; Liu, F.; Yao, Y.; Li, P.; Wang, D.; Wang, Z.; Chen, Y.; et al. Drug Release from Phase-Changeable Nanodroplets Triggered by Low-Intensity Focused Ultrasound. Theranostics 2018, 8, 1327–1339. [Google Scholar] [CrossRef]
- Zakeri-Milani, P.; Loveymi, B.D.; Jelvehgari, M.; Valizadeh, H. The Characteristics and Improved Intestinal Permeability of Vancomycin PLGA-Nanoparticles as Colloidal Drug Delivery System. Colloids Surf. B 2013, 103, 174–181. [Google Scholar] [CrossRef]
- Tefas, L.R.; Tomuţă, I.; Achim, M.; Vlase, L. Development and Optimization of Quercetin-Loaded PLGA Nanoparticles by Experimental Design. Clujul Med. 2015, 88, 214. [Google Scholar] [CrossRef] [Green Version]
- Mehta, N.; Sahu, S.P.; Shaik, S.; Devireddy, R.; Gartia, M.R. Dark-Field Hyperspectral Imaging for Label Free Detection of Nano-Bio-Materials. WIREs Nanomed. Nanobiotech. 2021, 13, e1661. [Google Scholar] [CrossRef]
- Jaidev, L.R.; Krishnan, U.M.; Sethuraman, S. Gemcitabine Loaded Biodegradable PLGA Nanospheres for in Vitro Pancreatic Cancer Therapy. Mater. Sci. Eng. C 2015, 47, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Jain, A.; Jain, S.K. In Vitro Release Kinetics Model Fitting of Liposomes: An Insight. Chem. Phys. Lipids 2016, 201, 28–40. [Google Scholar] [CrossRef]
- Aguzzi, C.; Cerezo, P.; Salcedo, I.; Sánchez, R.; Viseras, C. Mathematical Models Describing Drug Release from Biopolymeric Delivery Systems. Mater. Technol. 2010, 25, 205–211. [Google Scholar] [CrossRef]
- Gholizadeh, S.; Kamps Jan, A.A.M.; Hennink, W.E.; Kok, R.J. PLGA-PEG Nanoparticles for Targeted Delivery of the MTOR/PI3kinase Inhibitor Dactolisib to Inflamed Endothelium. Int. J. Pharm. 2018, 548, 747–758. [Google Scholar] [CrossRef] [PubMed]
- Sriamornsak, P.; Nunthanid, J.; Luangtana-anan, M.; Puttipipatkhachorn, S. Alginate-Based Pellets Prepared by Extrusion/Spheronization: A Preliminary Study on the Effect of Additive in Granulating Liquid. Eur. J. Pharm. Biopharm. 2007, 67, 227–235. [Google Scholar] [CrossRef] [PubMed]
- Mahapatro, A.; Singh, D.K. Biodegradable Nanoparticles Are Excellent Vehicle for Site Directed In-Vivo Delivery of Drugs and Vaccines. J. Nanobio. 2011, 9, 55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kapoor, D.N.; Bhatia, A.; Kaur, R.; Sharma, R.; Kaur, G.; Dhawan, S. PLGA: A Unique Polymer for Drug Delivery. Ther. Deliv. 2015, 6, 41–58. [Google Scholar] [CrossRef] [PubMed]
- Spreen, H.; Behrens, M.; Mulac, D.; Humpf, H.-U.; Langer, K. Identification of Main Influencing Factors on the Protein Corona Composition of PLGA and PLA Nanoparticles. Eur. J. Pharm. Biopharm. 2021, 163, 212–222. [Google Scholar] [CrossRef] [PubMed]
- Göppert, T.M.; Müller, R.H. Adsorption Kinetics of Plasma Proteins on Solid Lipid Nanoparticles for Drug Targeting. Int. J. Pharm. 2005, 302, 172–186. [Google Scholar] [CrossRef]
- Gossmann, R.; Fahrländer, E.; Hummel, M.; Mulac, D.; Brockmeyer, J.; Langer, K. Comparative Examination of Adsorption of Serum Proteins on HSA- and PLGA-Based Nanoparticles Using SDS–PAGE and LC–MS. Eur. J. Pharm. Biopharm. 2015, 93, 80–87. [Google Scholar] [CrossRef]
- Partikel, K.; Korte, R.; Stein, N.C.; Mulac, D.; Herrmann, F.C.; Humpf, H.-U.; Langer, K. Effect of Nanoparticle Size and PEGylation on the Protein Corona of PLGA Nanoparticles. Eur. J. Pharm. Biopharm. 2019, 141, 70–80. [Google Scholar] [CrossRef] [PubMed]
- Partikel, K.; Korte, R.; Mulac, D.; Humpf, H.-U.; Langer, K. Serum Type and Concentration Both Affect the Protein-Corona Composition of PLGA Nanoparticles. Beilstein J. Nanotech. 2019, 10, 1002–1015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ndumiso, M.; Buchtová, N.; Husselmann, L.; Mohamed, G.; Klein, A.; Aucamp, M.; Canevet, D.; d’Souza, S.; Maphasa, R.E.; Boury, F. Comparative Whole Corona Fingerprinting and Protein Adsorption Thermodynamics of PLGA and PCL Nanoparticles in Human Serum. Colloids Surf. B 2020, 188, 110816. [Google Scholar] [CrossRef]
- Ravi Kumar, M.N.V.; Bakowsky, U.; Lehr, C.M. Preparation and Characterization of Cationic PLGA Nanospheres as DNA Carriers. Biomaterials 2004, 25, 1771–1777. [Google Scholar] [CrossRef] [PubMed]
- Bainor, A.; Chang, L.; McQuade, T.J.; Webb, B.; Gestwicki, J.E. Bicinchoninic Acid (BCA) Assay in Low Volume. Anal. Biochem. 2011, 410, 310–312. [Google Scholar] [CrossRef] [PubMed]
- Pillai, G.J.; Greeshma, M.M.; Menon, D. Impact of Poly(Lactic-Co-Glycolic Acid) Nanoparticle Surface Charge on Protein, Cellular and Haematological Interactions. Colloids Surf. B 2015, 136, 1058–1066. [Google Scholar] [CrossRef] [PubMed]
- Salatin, S.; Maleki Dizaj, S.; Yari Khosroushahi, A. Effect of the Surface Modification, Size, and Shape on Cellular Uptake of Nanoparticles. Cell Biol. Int. 2015, 39, 881–890. [Google Scholar] [CrossRef]
- Dobrovolskaia, M.A.; Neun, B.W.; Man, S.; Ye, X.; Hansen, M.; Patri, A.K.; Crist, R.M.; McNeil, S.E. Protein Corona Composition Does Not Accurately Predict Hematocompatibility of Colloidal Gold Nanoparticles. Nanomedicine 2014, 10, 1453–1463. [Google Scholar] [CrossRef] [Green Version]
- Patil, S.; Sandberg, A.; Heckert, E.; Self, W.; Seal, S. Protein Adsorption and Cellular Uptake of Cerium Oxide Nanoparticles as a Function of Zeta Potential. Biomaterials 2007, 28, 4600–4607. [Google Scholar] [CrossRef] [Green Version]
- Weber, C.; Morsbach, S.; Landfester, K. Possibilities and Limitations of Different Separation Techniques for the Analysis of the Protein Corona. Angew. Chem. Int. Ed. 2019, 58, 12787–12794. [Google Scholar] [CrossRef]
- Ahsan, S.M.; Rao, C.M.; Ahmad, M.F. Nanoparticle-Protein Interaction: The Significance and Role of Protein Corona. In Cellular and Molecular Toxicology of Nanoparticles; Saquib, Q., Faisal, M., Al-Khedhairy, A.A., Alatar, A.A., Eds.; Springer International Publishing: Cham, Switzerland, 2018; pp. 175–198. ISBN 978-3-319-72041-8. [Google Scholar]
- Zhang, Y.; Xu, Y.-Y.; Sun, W.-J.; Zhang, M.-H.; Zheng, Y.-F.; Shen, H.-M.; Yang, J.; Zhu, X.-Q. FBS or BSA Inhibits EGCG Induced Cell Death through Covalent Binding and the Reduction of Intracellular ROS Production. BioMed Res. Int. 2016, 2016, 5013409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meloun, B.; Morávek, L.; Kostka, V. Complete Amino Acid Sequence of Human Serum Albumin. FEBS Lett. 1975, 58, 134–137. [Google Scholar] [CrossRef] [Green Version]
- Szekeres, G.P.; Kneipp, J. Different Binding Sites of Serum Albumins in the Protein Corona of Gold Nanoparticles. Analyst 2018, 143, 6061–6068. [Google Scholar] [CrossRef] [PubMed]
- Settanni, G.; Zhou, J.; Suo, T.; Schöttler, S.; Landfester, K.; Schmid, F.; Mailänder, V. Protein Corona Composition of Poly(Ethylene Glycol)- and Poly(Phosphoester)-Coated Nanoparticles Correlates Strongly with the Amino Acid Composition of the Protein Surface. Nanoscale 2017, 9, 2138–2144. [Google Scholar] [CrossRef] [Green Version]
- Naoi, M.; Wu, Y.; Shamoto-Nagai, M.; Maruyama, W. Mitochondria in Neuroprotection by Phytochemicals: Bioactive Polyphenols Modulate Mitochondrial Apoptosis System, Function and Structure. Int. J. Mol. Sci. 2019, 20, 2451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, A.; Jahan, S.; Alshahrani, S.; Alshehri, B.M.; Sameer, A.S.; Arafah, A.; Ahmad, A.; Rehman, M.U. Chapter 21—Phytotherapeutic Agents for Neurodegenerative Disorders: A Neuropharmacological Review. In Phytomedicine; Bhat, R.A., Hakeem, K.R., Dervash, M.A., Eds.; Academic Press: Cambridge, MA, USA, 2021; pp. 581–620. ISBN 978-0-12-824109-7. [Google Scholar]
- Gaikwad, P.; Barik, A.; Priyadarsini, K.I.; Rao, B.S.M. Antioxidant Activities of Phenols in Different Solvents Using DPPH Assay. Res. Chem. Intermed. 2010, 36, 1065–1072. [Google Scholar] [CrossRef]
- Djiokeng Paka, G.; Doggui, S.; Zaghmi, A.; Safar, R.; Dao, L.; Reisch, A.; Klymchenko, A.; Roullin, V.G.; Joubert, O.; Ramassamy, C. Neuronal Uptake and Neuroprotective Properties of Curcumin-Loaded Nanoparticles on SK-N-SH Cell Line: Role of Poly (Lactide-Co-Glycolide) Polymeric Matrix Composition. Mol. Pharm. 2016, 13, 391–403. [Google Scholar] [CrossRef]
- Catterall, K.; Robertson, D.; Hudson, S.; Teasdale, P.R.; Welsh, D.T.; John, R. A Sensitive, Rapid Ferricyanide-Mediated Toxicity Bioassay Developed Using Escherichia Coli. Talanta 2010, 82, 751–757. [Google Scholar] [CrossRef]
- Tacchini, M.; Echeverria Guevara, M.P.; Grandini, A.; Maresca, I.; Radice, M.; Angiolella, L.; Guerrini, A. Ocimum Campechianum Mill. from Amazonian Ecuador: Chemical Composition and Biological Activities of Extracts and Their Main Constituents (Eugenol and Rosmarinic Acid). Molecules 2021, 26, 84. [Google Scholar] [CrossRef]
- Mosmann, T. Rapid Colorimetric Assay for Cellular Growth and Survival: Application to Proliferation and Cytotoxicity Assays. J. Immunol. Meth. 1983, 65, 55–63. [Google Scholar] [CrossRef]
- Fornaguera, C.; Feiner-Gracia, N.; Calderó, G.; García-Celma, M.J.; Solans, C. Galantamine-Loaded PLGA Nanoparticles, from Nano-Emulsion Templating, as Novel Advanced Drug Delivery Systems to Treat Neurodegenerative Diseases. Nanoscale 2015, 7, 12076–12084. [Google Scholar] [CrossRef] [PubMed]
- Chiu, H.I.; Samad, N.A.; Fang, L.; Lim, V. Cytotoxicity of Targeted PLGA Nanoparticles: A Systematic Review. RSC Adv. 2021, 11, 9433–9449. [Google Scholar] [CrossRef] [PubMed]
- Mishra, R.K.; Ahmad, A.; Vyawahare, A.; Alam, P.; Khan, T.H.; Khan, R. Biological Effects of Formation of Protein Corona onto Nanoparticles. Int. J. Biol. Macromol. 2021, 175, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Macey, M.G. Principles of Flow Cytometry. In Flow Cytometry: Principles and Applications; Macey, M.G., Ed.; Humana Press: Totowa, NJ, USA, 2007; pp. 1–15. ISBN 978-1-59745-451-3. [Google Scholar]





| Entry | Nanoparticle (NP) | RA (mg) | Organic (Oil) Phase (EtOAc:EtOH) | Method 2 | EE% 3 | Average Diameter 4 (nm) | PDI | ζ-Potential 5 (mV) |
|---|---|---|---|---|---|---|---|---|
| 1 | PLGA | 0 | 100:0 | A | - | 46.6 ± 0.68 | 0.29 | −18.0 ± 6.4 |
| 2 | RA-loaded PLGA | 1.2 | 100:0 | B | 92.2 ± 4.62 | 74.7 ± 0.17 | 0.30 | −21.9 ± 1.26 |
| 3 | RA-loaded PLGA | 1.6 | 100:0 | B | 80.7 ± 7.15 | 70.9 ± 2.31 | 0.29 | −36.8 ± 1.96 |
| 4 | RA-loaded PLGA | 1.6 | 90:10 | A | 64.5 ± 4.80 | 167.9 ± 3.25 | 0.21 | −35.5 ± 6.0 |
| Sample (NPs) | Incubation Time (Hours) | FBS (10%) | Average Diameter (nm) | PDI | ζ-Potential (mV) | Protein Corona Concentration (µg·mL−1) |
|---|---|---|---|---|---|---|
| PLGA 2 | 1 | No | 41.8 ± 1.25 | 0.27 | −12.7 ± 0.62 | - |
| PLGA 2 | 5 | No | 43.4 ± 0.30 | 0.40 | −13.1 ± 0.83 | - |
| PLGA_PC | 1 | Yes | 94.2 ± 2.49 | 0.22 | −25.5 ± 4.73 | 103.1 ± 6.05 |
| PLGA_PC | 5 | Yes | 95.3 ± 4.45 | 0.21 | −25.1 ± 3.61 | 141.8 ± 11.4 |
| RA-loaded PLGA 3 | 1 | No | 86.8 ± 1.02 | 0.28 | −19.0 ± 1.27 | - |
| RA-loaded PLGA 3 | 5 | No | 87.5 ± 1.49 | 0.27 | −13.1 ± 0.69 | - |
| RA-loaded PLGA_PC | 1 | Yes | 129.6 ± 4.85 | 0.18 | −33.0 ± 2.65 | 188.1 ± 1.75 |
| RA-loaded PLGA_PC | 5 | Yes | 137.3 ± 4.76 | 0.17 | −20.4 ± 1.38 | 118.7 ± 8.33 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
García-Melero, J.; López-Mitjavila, J.-J.; García-Celma, M.J.; Rodriguez-Abreu, C.; Grijalvo, S. Rosmarinic Acid-Loaded Polymeric Nanoparticles Prepared by Low-Energy Nano-Emulsion Templating: Formulation, Biophysical Characterization, and In Vitro Studies. Materials 2022, 15, 4572. https://doi.org/10.3390/ma15134572
García-Melero J, López-Mitjavila J-J, García-Celma MJ, Rodriguez-Abreu C, Grijalvo S. Rosmarinic Acid-Loaded Polymeric Nanoparticles Prepared by Low-Energy Nano-Emulsion Templating: Formulation, Biophysical Characterization, and In Vitro Studies. Materials. 2022; 15(13):4572. https://doi.org/10.3390/ma15134572
Chicago/Turabian StyleGarcía-Melero, Jessica, Joan-Josep López-Mitjavila, María José García-Celma, Carlos Rodriguez-Abreu, and Santiago Grijalvo. 2022. "Rosmarinic Acid-Loaded Polymeric Nanoparticles Prepared by Low-Energy Nano-Emulsion Templating: Formulation, Biophysical Characterization, and In Vitro Studies" Materials 15, no. 13: 4572. https://doi.org/10.3390/ma15134572






