Laser-Induced Transferred Antibacterial Nanoparticles for Mixed-Species Bacteria Biofilm Inactivation
Abstract
:1. Introduction
2. Materials and Methods
3. Results and Discussions
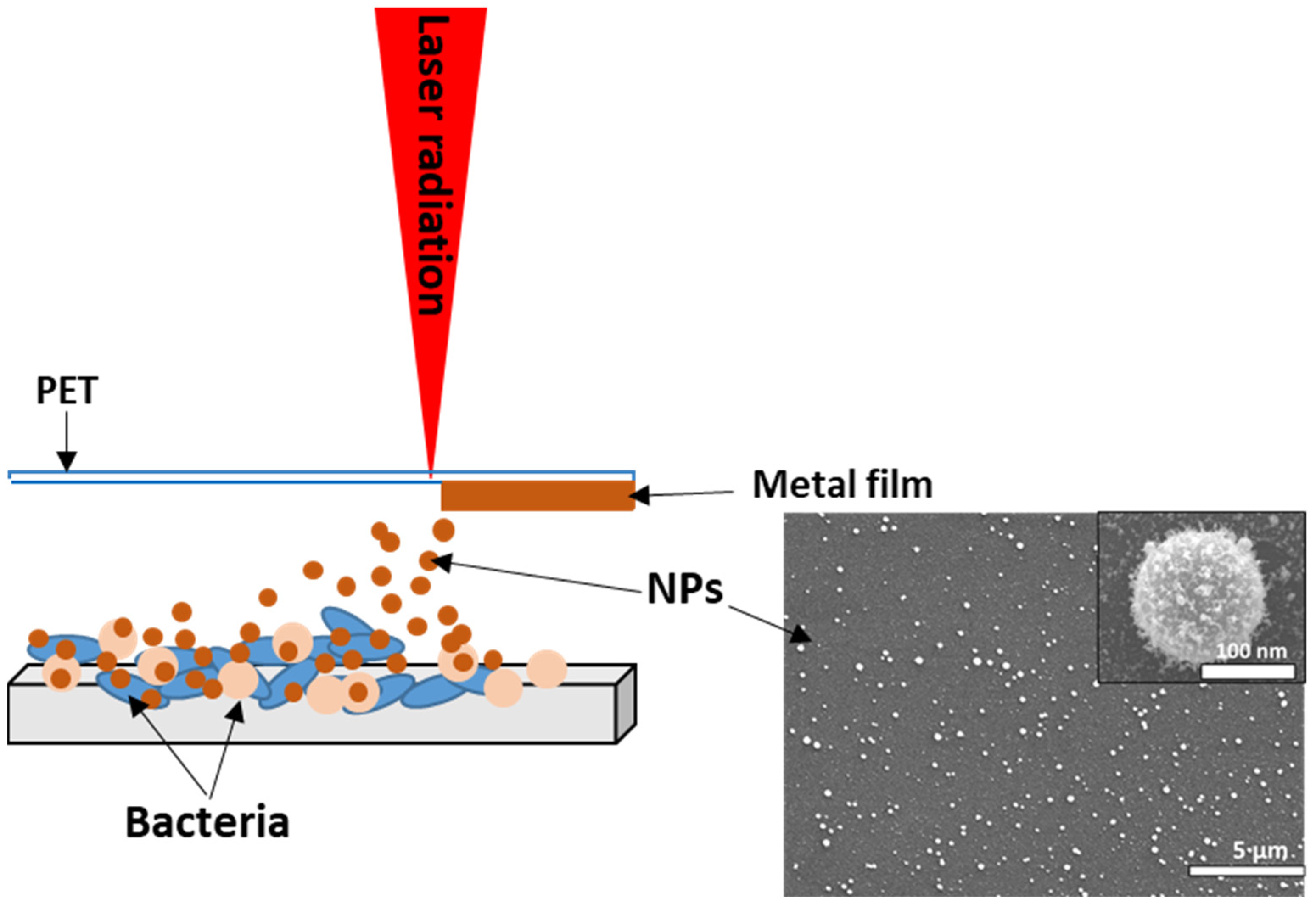
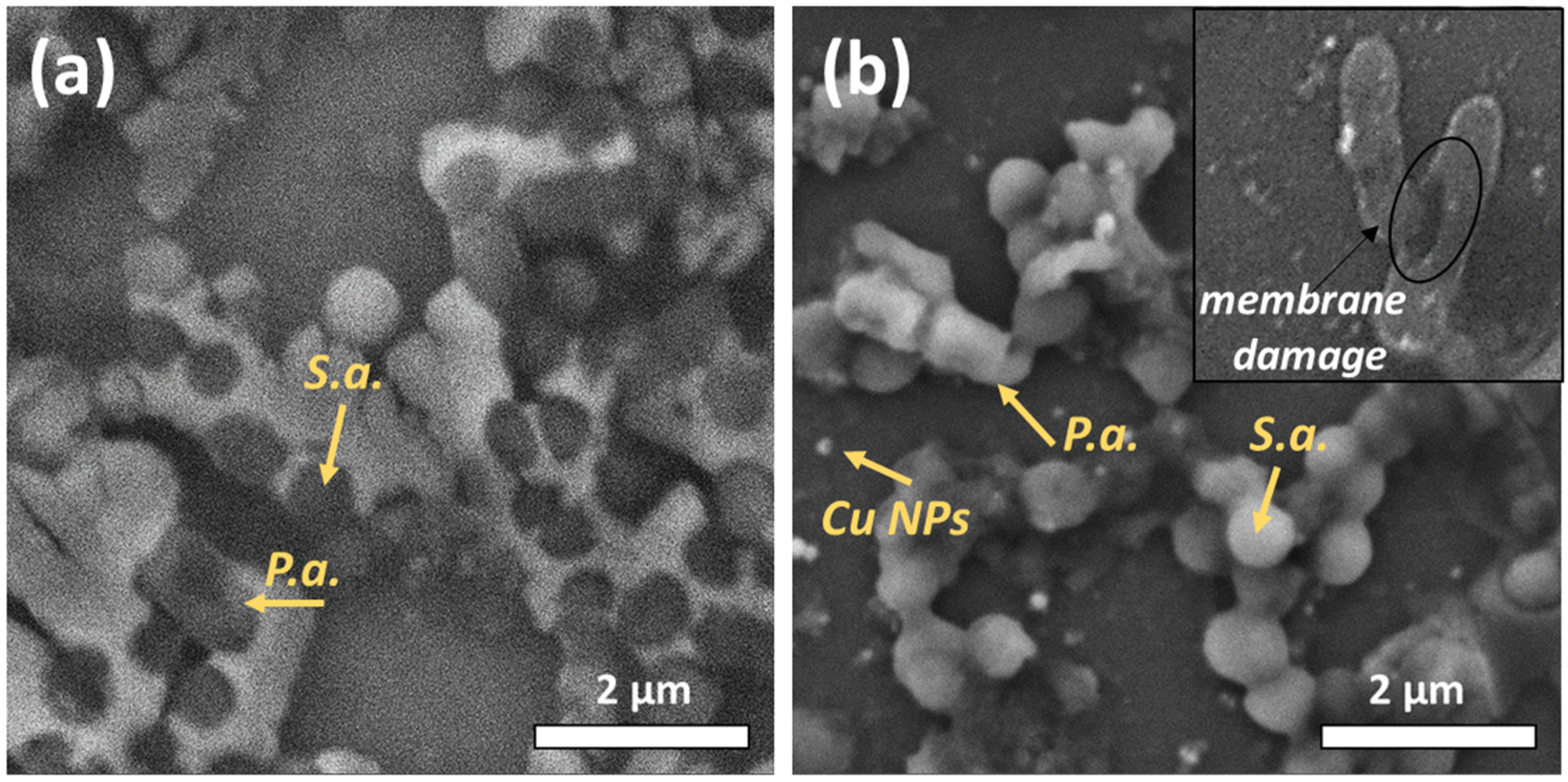
3.1. SEM Characterization
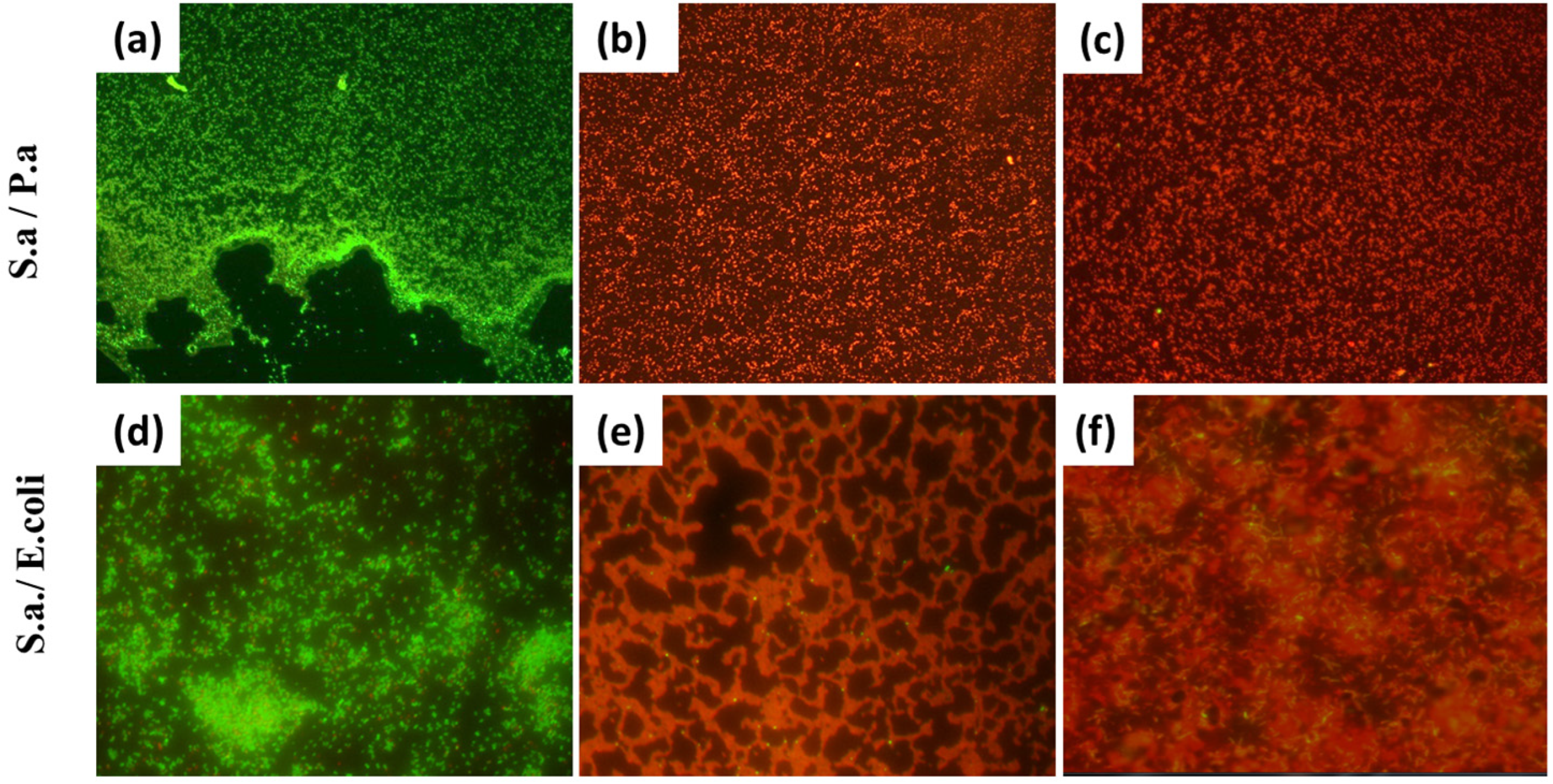
3.2. Investigation of the Antibacterial Effect of Nanoparticles on Mixed-Species Biofilms
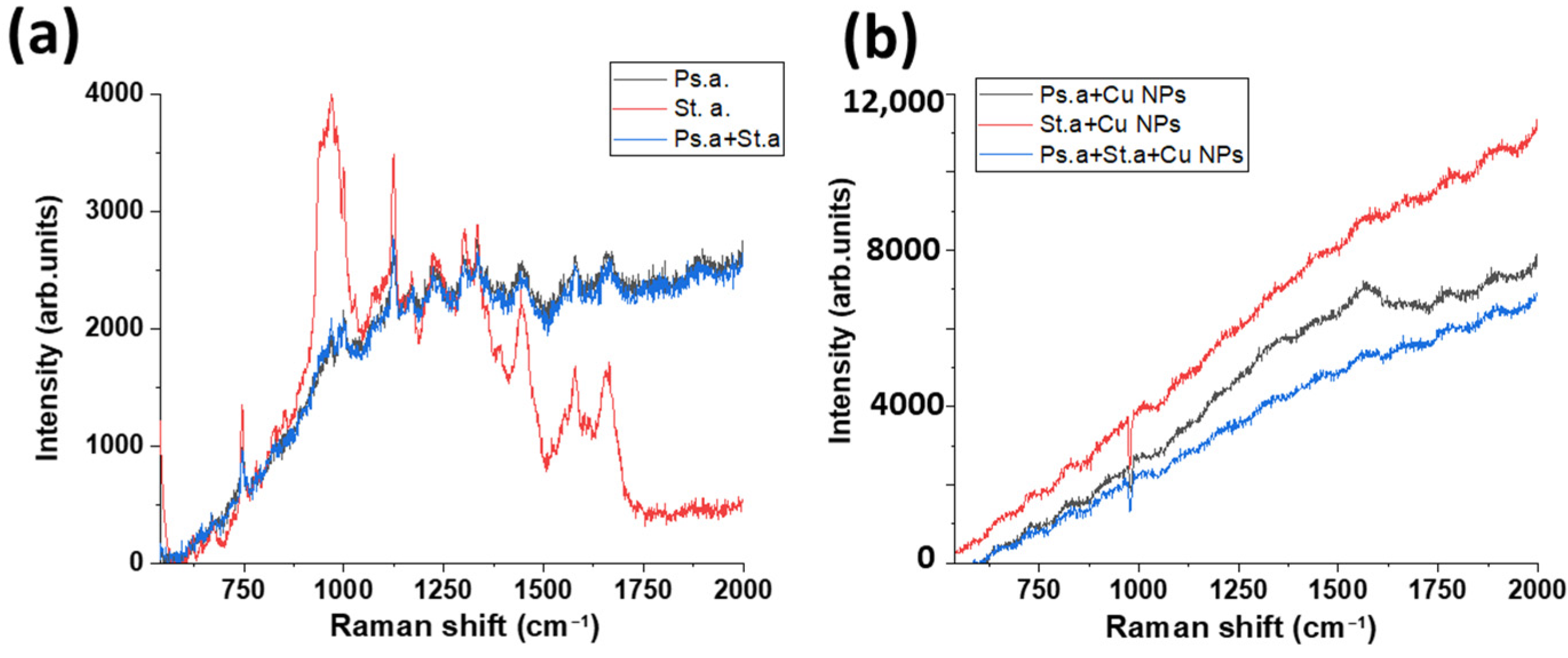
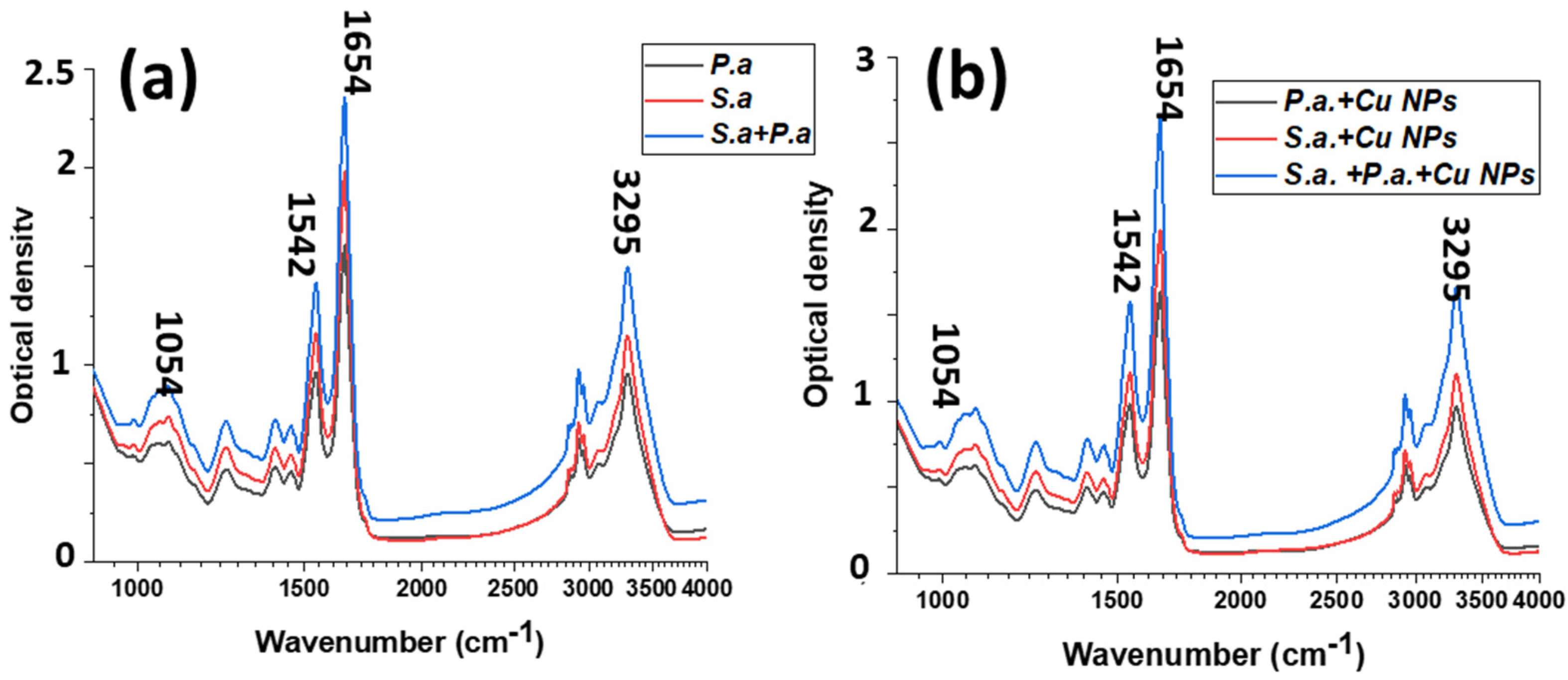
3.3. Raman and FTIR Spectroscopy of Mixed-Species Bacteria Staphylococcus aureus and Pseudomonas aeruginosa
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- López, D.; Vlamakis, H.; Kolter, R. Biofilms. Cold Spring Harb. Perspect. Biol. 2010, 2, a000398. [Google Scholar] [CrossRef] [PubMed]
- Thi, M.T.T.; Wibowo, D.; Rehm, B.H.A. Pseudomonas aeruginosa biofilms. Int. J. Mol. Sci. 2020, 21, 8671. [Google Scholar] [CrossRef] [PubMed]
- Hatt, J.K.; Rather, P.N. Role of bacterial biofilms in urinary tract infections. Bact. Biofilms 2008, 322, 163–192. [Google Scholar] [CrossRef]
- Costerton, J.W.; Geesey, G.G.; Cheng, K.J. How bacteria stick. Sci. Am. 1978, 238, 86–95. [Google Scholar] [CrossRef]
- Jamal, M.; Ahmad, W.; Andleeb, S.; Jalil, F.; Imran, M.; Nawaz, M.A.; Hussain, T.; Ali, M.; Rafiq, M.; Kamil, M.A. Bacterial biofilm and associated infections. J. Chin. Med. Assoc. 2018, 81, 7–11. [Google Scholar] [CrossRef]
- Ilyina, T.S.; Romanova, Y.M. Bacterial biofilms: Their role in chronical infection processes and the means to combat them. Mol. Gen. Microbiol. Virol. 2021, 39, 14–24. [Google Scholar] [CrossRef]
- Rajpaul, K. Biofilm in wound care. Br. J. Community Nurs. 2015, 20, 6–11. [Google Scholar] [CrossRef]
- Yung, D.B.Y.; Sircombe, K.J.; Pletzer, D. Friends or enemies? The complicated relationship between Pseudomonas aeruginosa and Staphylococcus aureus. Mol. Microbiol. 2021, 116, 1–15. [Google Scholar] [CrossRef]
- Alves, P.M.; Al-Badi, E.; Withycombe, C.; Jones, P.M.; Purdy, K.J.; Maddocks, S.E. Interaction between Staphylococcus aureus and Pseudomonas aeruginosa is beneficial for colonisation and pathogenicity in a mixed biofilm. Pathog. Dis. 2018, 76, fty003. [Google Scholar] [CrossRef] [Green Version]
- Nastulyavichus, A.; Khaertdinova, L.; Tolordava, E.; Yushina, Y.; Ionin, A.; Semenova, A.; Kudryashov, S. Additive Nanosecond Laser-Induced Forward Transfer of High Antibacterial Metal Nanoparticle Dose onto Foodborne Bacterial Biofilms. Micromachines 2022, 13, 2170. [Google Scholar] [CrossRef]
- Nastulyavichus, A.; Tolordava, E.; Rudenko, A.; Zazymkina, D.; Shakhov, P.; Busleev, N.; Romanova, Y.; Ionin, A.; Kudryashov, S. In vitro destruction of pathogenic bacterial biofilms by bactericidal metallic nanoparticles via laser-induced forward transfer. Nanomaterials 2020, 10, 2259. [Google Scholar] [CrossRef]
- Nastulyavichus, A.; Kudryashov, S.; Smirnov, N.; Saraeva, I.; Rudenko, A.; Tolordava, E.; Zayarny, D. Antibacterial coatings of Se and Si nanoparticles. Appl. Surf. Sci. 2019, 469, 220–225. [Google Scholar] [CrossRef]
- Qayyum, S.; Khan, A.U. Nanoparticles vs. biofilms: A battle against another paradigm of antibiotic resistance. MedChemComm 2016, 7, 1479–1498. [Google Scholar] [CrossRef]
- Tan, Y.; Ma, S.; Leonhard, M.; Moser, D.; Haselmann, G.M.; Wang, J.; Dominik, E.; Stickler, B. Enhancing antibiofilm activity with functional chitosan nanoparticles targeting biofilm cells and biofilm matrix. Carbohydr. Polym. 2018, 200, 35–42. [Google Scholar] [CrossRef]
- Rao, Y.; Shang, W.; Yang, Y.; Zhou, R.; Rao, X. Fighting mixed-species microbial biofilms with cold atmospheric plasma. Front. Microbiol. 2020, 11, 1000. [Google Scholar] [CrossRef]
- Pankaj, S.K.; Wan, Z.; Colonna, W.; Keener, K.M. Effect of high voltage atmospheric cold plasma on white grape juice quality. J. Sci. Food Agric. 2017, 97, 4016–4021. [Google Scholar] [CrossRef]
- Govaert, M.; Smet, C.L.; Walsh, J.; Van Impe, J.F. Influence of plasma characteristics on the inactivation mechanism of cold atmospheric plasma (CAP) for Listeria monocytogenes and Salmonella typhimurium biofilms. Appl. Sci. 2020, 10, 3198. [Google Scholar] [CrossRef]
- Bonvicini, F.; Belluti, F.; Bisi, A.; Gobbi, S.; Manet, I.; Gentilomi, G.A. Improved eradication efficacy of a combination of newly identified antimicrobial agents in C. albicans and S. aureus mixed-species biofilm. Res. Microbiol. 2021, 172, 103873. [Google Scholar] [CrossRef]
- Mgomi, F.C.; Yuan, L.; Chen, C.W.; Zhang, Y.S.; Yang, Z.Q. Bacteriophages: A weapon against mixed-species biofilms in the food processing environment. J. Appl. Microbiol. 2022, 133, 2107–2121. [Google Scholar] [CrossRef]
- Milho, C.; Silva, M.D.; Alves, D.; Oliveira, H.; Sousa, C.; Pastrana, L.M.; Azeredo, J.; Sillankorva, S. Escherichia coli and Salmonella enteritidis dual-species biofilms: Interspecies interactions and antibiofilm efficacy of phages. Sci. Rep. 2019, 9, 18183. [Google Scholar] [CrossRef] [Green Version]
- Jahid, I.K.; Han, N.R.; Srey, S.; Ha, S.D. Competitive interactions inside mixed-culture biofilms of Salmonella typhimurium and cultivable indigenous microorganisms on lettuce enhance microbial resistance of their sessile cells to ultraviolet C (UV-C) irradiation. Food Res. Int. 2014, 55, 445–4554. [Google Scholar] [CrossRef]
- Pourhajibagher, M.; Rahimi-Esboei, B.; Ahmadi, H.; Bahador, A. The anti-biofilm capability of nano-emodin-mediated sonodynamic therapy on multi-species biofilms produced by burn wound bacterial strains. Photodiagnosis Photodyn. Ther. 2021, 34, 102288. [Google Scholar] [CrossRef] [PubMed]
- Roy, R.; Tiwari, M.; Donelli, G.; Tiwari, V. Strategies for combating bacterial biofilms: A focus on anti-biofilm agents and their mechanisms of action. Virulence 2018, 9, 522–554. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Hu, C.; Shao, L. The antimicrobial activity of nanoparticles: Present situation and prospects for the future. Int. J. Nanomedicine 2017, 12, 1227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khashan, K.S.; Sulaiman, G.M.; Abdulameer, F.A. Synthesis and antibacterial activity of CuO nanoparticles suspension induced by laser ablation in liquid. Arab. J. Sci. Eng. 2016, 41, 301–310. [Google Scholar] [CrossRef]
- Raffi, M.; Mehrwan, S.; Bhatti, T.M.; Akhter, J.I.; Hameed, A.; Yawar, W. Investigations into the antibacterial behavior of copper nanoparticles against Escherichia coli. Ann. Microbiol. 2010, 60, 75–80. [Google Scholar] [CrossRef]
- Ghotaslou, R.; Bahari, Z.; Aliloo, A.; Gholizadeh, P.; Eshlaghi, B.S. The in vitro effects of silver nanoparticles on bacterial biofilms. J. Microbiol. Biotechnol. Food Sci. 2021, 6, 1077–1080. [Google Scholar] [CrossRef] [Green Version]
- Hosnedlova, B.; Kabanov, D.; Kepinska, M.; B Narayanan, V.H.; Parikesit, A.A.; Fernandez, C.; Bjorklund, G.; Nguen, H.V.; Farid, A.; Sochor, J.; et al. Effect of Biosynthesized Silver Nanoparticles on Bacterial Biofilm Changes in S. aureus and E. coli. Nanomaterials. 2022, 12, 2183. [Google Scholar] [CrossRef]
- Chong, W.X.; Lai, Y.X.; Choudhury, M.; Amalraj, F.D. Efficacy of incorporating silver nanoparticles into maxillofacial silicone against Staphylococcus aureus, Candida albicans, and polymicrobial biofilms. J. Prosthet. Dent. 2022, 128, 1114–1120. [Google Scholar] [CrossRef]
- Barros, J.; Grenho, L.; Fontenente, S.; Manuel, C.M.; Nunes, O.C.; Melo, L.F.; Monteiro, F.J.; Ferraz, M.P. Staphylococcus aureus and Escherichia coli dual-species biofilms on nanohydroxyapatite loaded with CHX or ZnO nanoparticles. J. Biomed. Mater. Res. A 2017, 105, 491–497. [Google Scholar] [CrossRef]
- Bhatia, E.; Banerjee, R. Hybrid silver–gold nanoparticles suppress drug resistant polymicrobial biofilm formation and intracellular infection. J. Mater. Chem. B 2020, 8, 4890–4898. [Google Scholar] [CrossRef]
- Tang, J.W.; Li, J.Q.; Yin, X.C.; Xu, W.W.; Pan, Y.C.; Liu, Q.H.; Gu, B.; Zhang, X.; Wang, L. Rapid discrimination of clinically important pathogens through machine learning analysis of surface enhanced Raman spectra. Front. Microbiol. 2022, 13, 1115. [Google Scholar] [CrossRef]
- Zarnowiec, P.; Lechowicz, L.; Czerwonka, G.; Kaca, W. Fourier transform infrared spectroscopy (FTIR) as a tool for the identification and differentiation of pathogenic bacteria. Curr. Med. Chem. 2015, 22, 1710–1718. [Google Scholar] [CrossRef]
- Vu, X.H.; Duong, T.T.T.; Pham, T.T.H.; Trinh, D.K.; Nguyen, X.H.; Dang, V.S. Synthesis and study of silver nanoparticles for antibacterial activity against Escherichia coli and Staphylococcus aureus. Adv. Nat. Sci. Nanosci. Nanotechnol. 2018, 9, 025019. [Google Scholar] [CrossRef]
- Saraeva, I.; Tolordava, E.; Sheligyna, S.; Nastulyavichus, A.; Khmelnitskii, R.; Pokryshkin, N.; Khmelenin, D.; Kudryashov, S.; Ionin, A.; Akhmatkhanov, A. FT-IR Analysis of P. aeruginosa Bacteria Inactivation by Femtosecond IR Laser Radiation. Int. J. Mol. Sci. 2023, 24, 5119. [Google Scholar] [CrossRef]



| Bacteria | Ag NPs | Cu NPs | Control |
|---|---|---|---|
| S. aureus | <1 | <1 | 4 × 106 |
| P. aeruginosa | <1 | <1 | 3 × 107 |
| P. aeruginosa + E. coli | <5 | <5 | 8 × 107 |
| S. aureus + E. coli | <5 | <5 | 2 × 107 |
| S. aureus + P. aeruginosa | <5 | <5 | 1 × 105 |
| Peak (cm−1) | Assignments |
|---|---|
| 745 | Ring breathing mode of DNA/ RNA bases thymine |
| 1120 | C–C and C–N assigned to adenine/thymine and stretching mode of amide III |
| 1158 | C-C, C-N stretching (protein) |
| 1307 | CH2 deformation in proteins and lipids |
| 1447 | CH2 deformation, CH3/CH2 twisting in carbohydrates, proteins and lipids |
| 1578 | Ring breathing modes in the DNA bases |
| 1653 | Stretching mode of amide I |
| Peak (cm−1) | Assignments |
|---|---|
| 1054 | P=O stretches of phosphodiesters |
| 1236 | P=O stretches of phosphodiesters |
| 1454 | CH3 asymmetric and symmetric deformation of proteins |
| 1545 | Amide II bands, referred to N-H deformation of amides associated with proteins |
| 1654 | Amide I band, C=O stretching vibrations of amides associated with proteins |
| 1800–1400 | Vibrational feature of group protein-associated |
| 2960 | C-H str (asym) in CH3 of fatty acids |
| 3290 | O-H stretch |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nastulyavichus, A.; Tolordava, E.; Kudryashov, S.; Khmelnitskii, R.; Ionin, A. Laser-Induced Transferred Antibacterial Nanoparticles for Mixed-Species Bacteria Biofilm Inactivation. Materials 2023, 16, 4309. https://doi.org/10.3390/ma16124309
Nastulyavichus A, Tolordava E, Kudryashov S, Khmelnitskii R, Ionin A. Laser-Induced Transferred Antibacterial Nanoparticles for Mixed-Species Bacteria Biofilm Inactivation. Materials. 2023; 16(12):4309. https://doi.org/10.3390/ma16124309
Chicago/Turabian StyleNastulyavichus, Alena, Eteri Tolordava, Sergey Kudryashov, Roman Khmelnitskii, and Andrey Ionin. 2023. "Laser-Induced Transferred Antibacterial Nanoparticles for Mixed-Species Bacteria Biofilm Inactivation" Materials 16, no. 12: 4309. https://doi.org/10.3390/ma16124309





