A Novel Approach for Automated Analysis of Cell Attachment and Spreading Based on Backscattered Electron Imaging by Scanning Electron Microscopy
Abstract
:1. Introduction
2. Results and Discussion
2.1.Tested materials
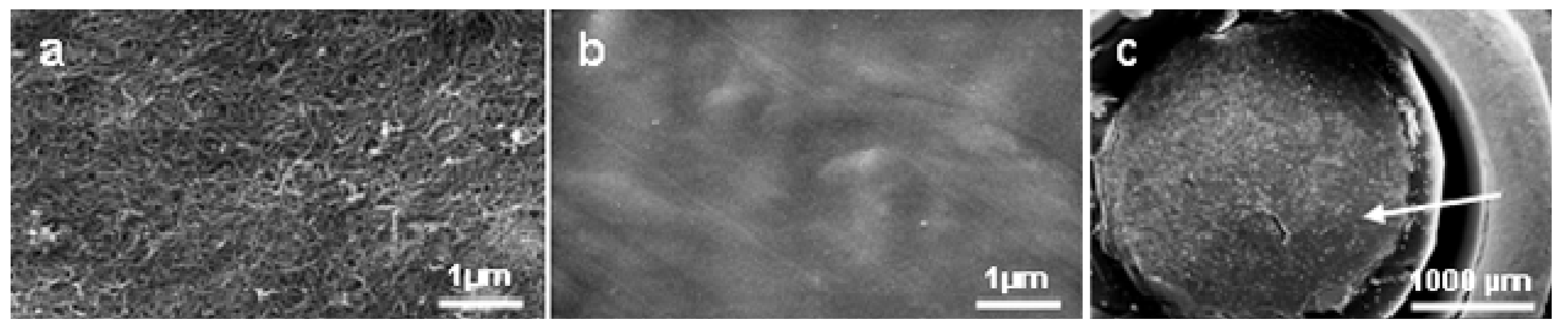
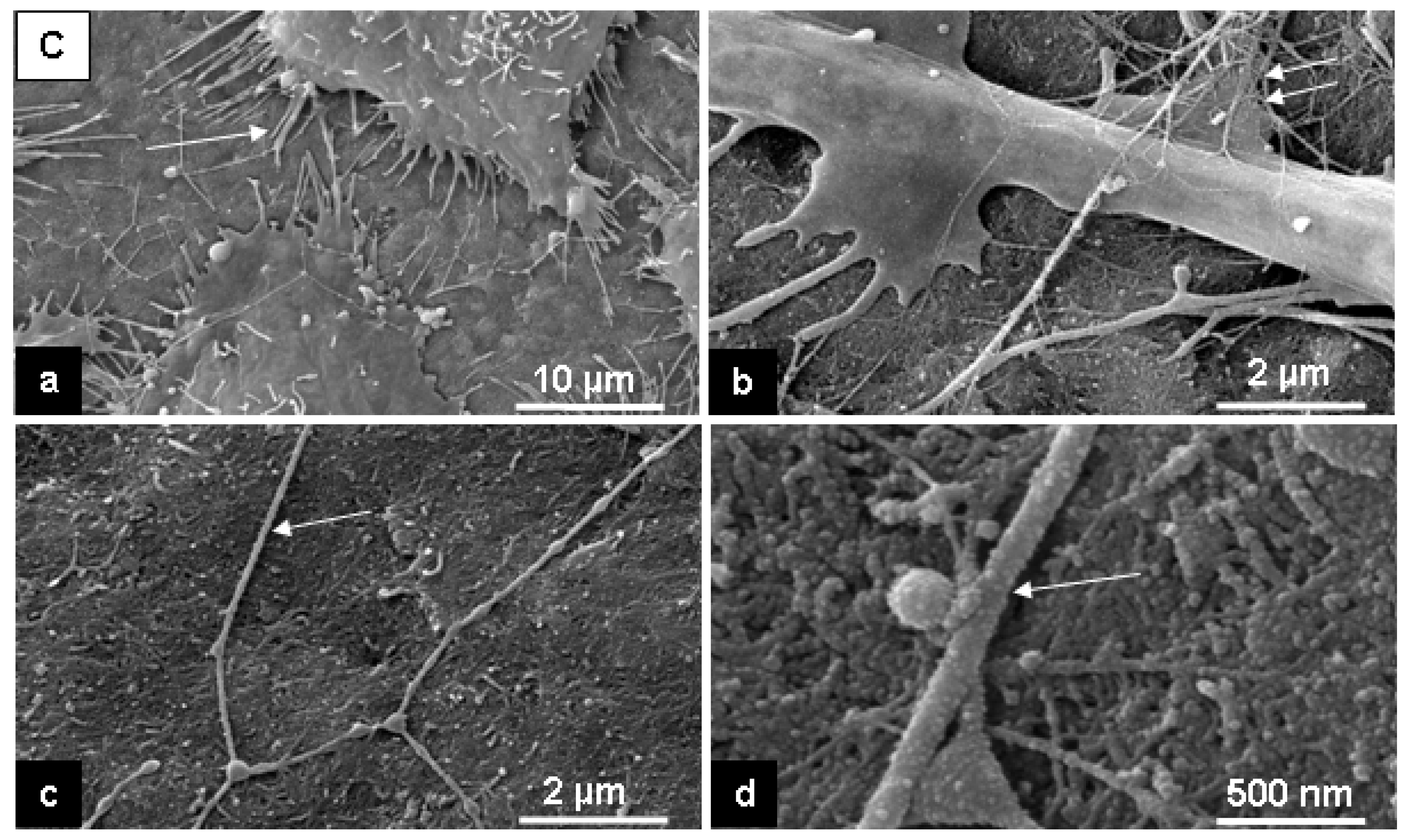
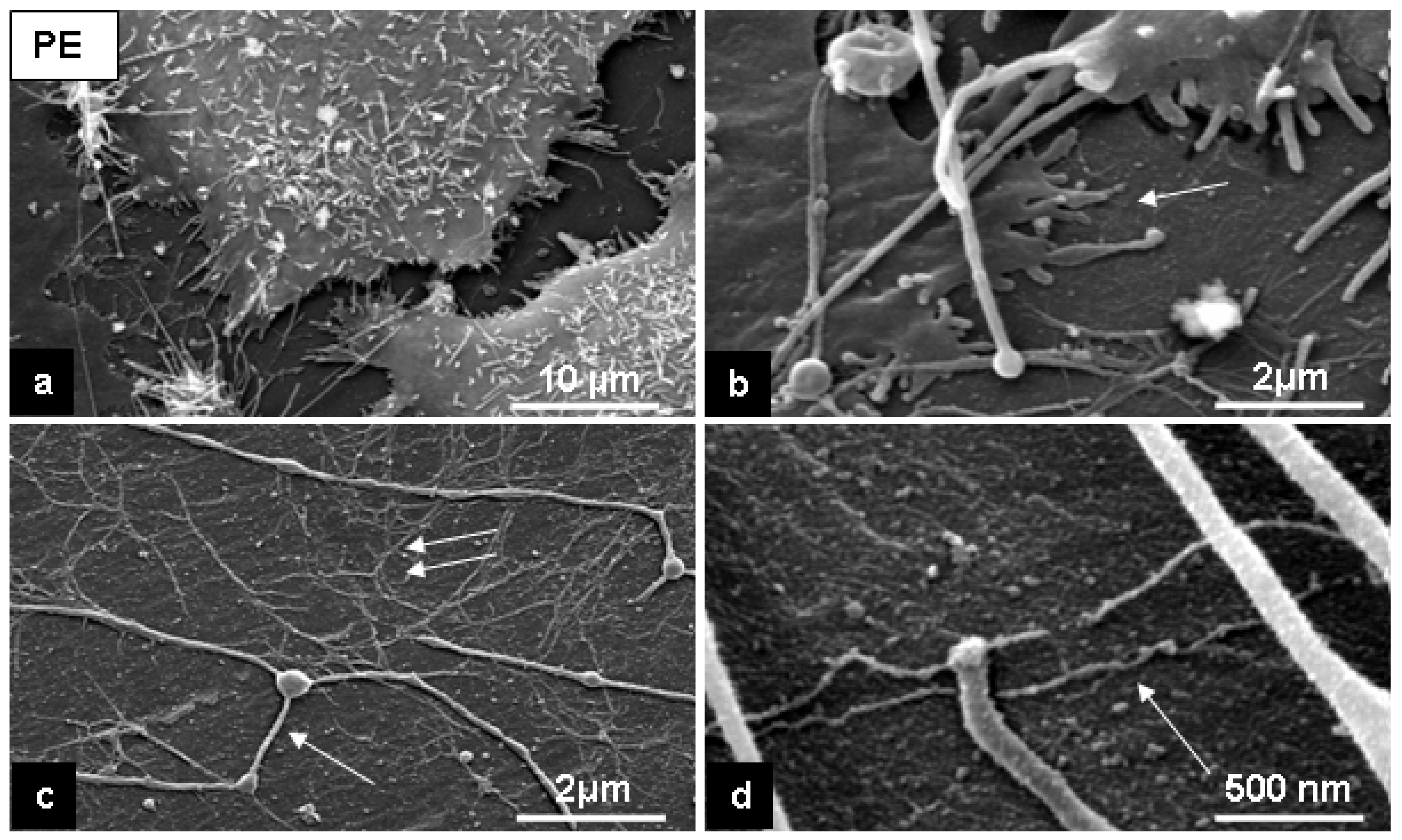
2.2. Cell response to chosen materials
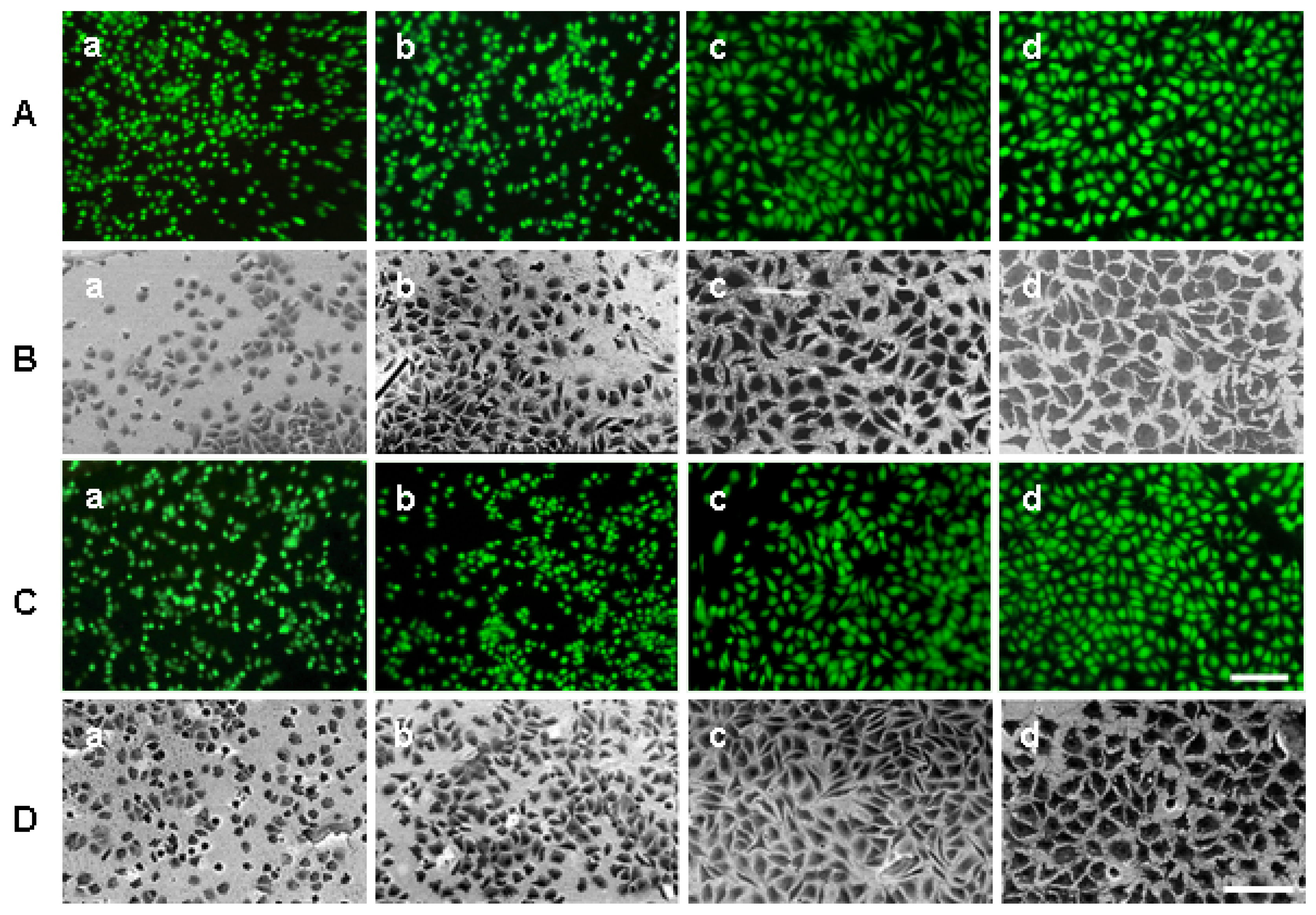

2.3. Quantification of cell attachment & spreading
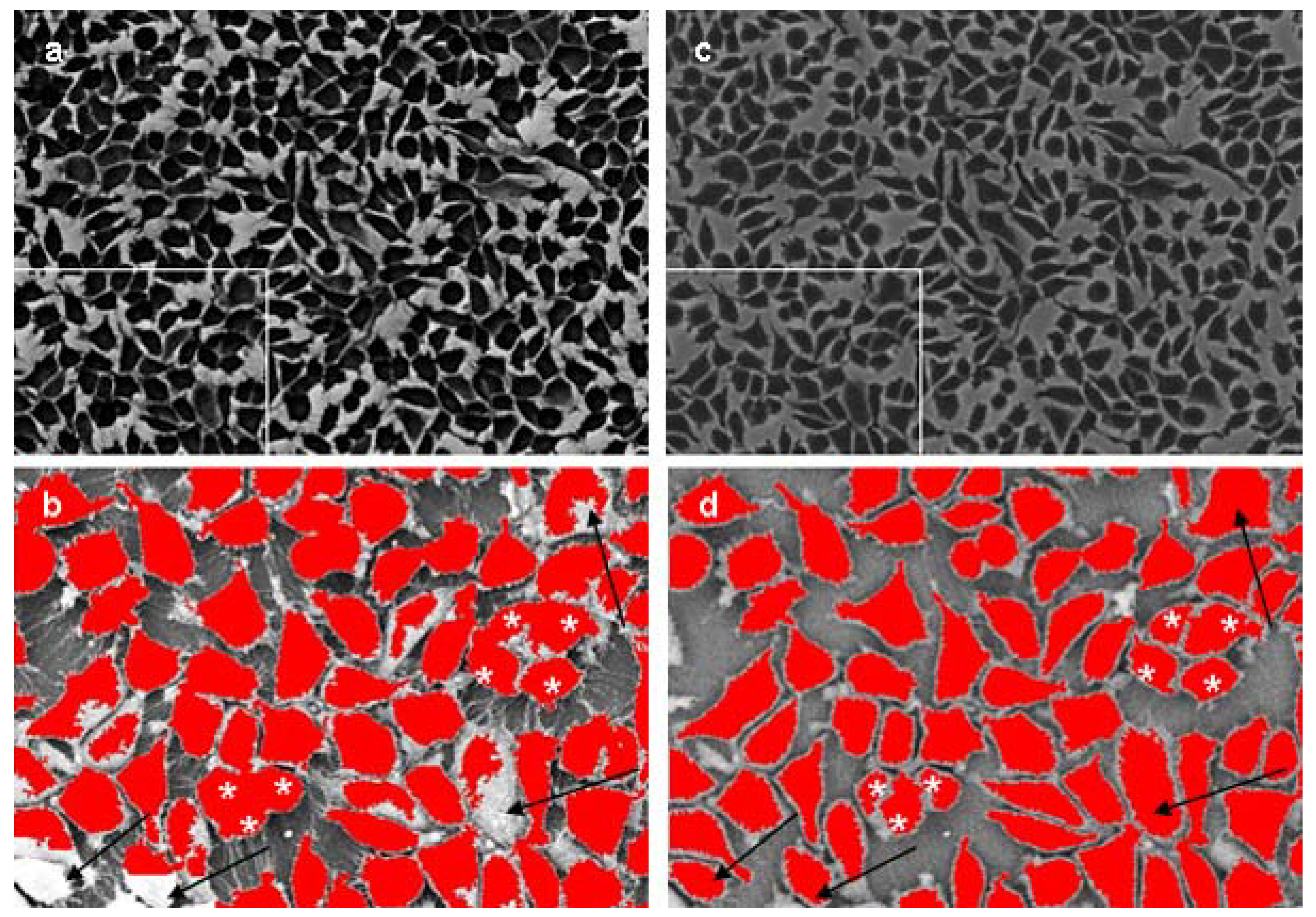
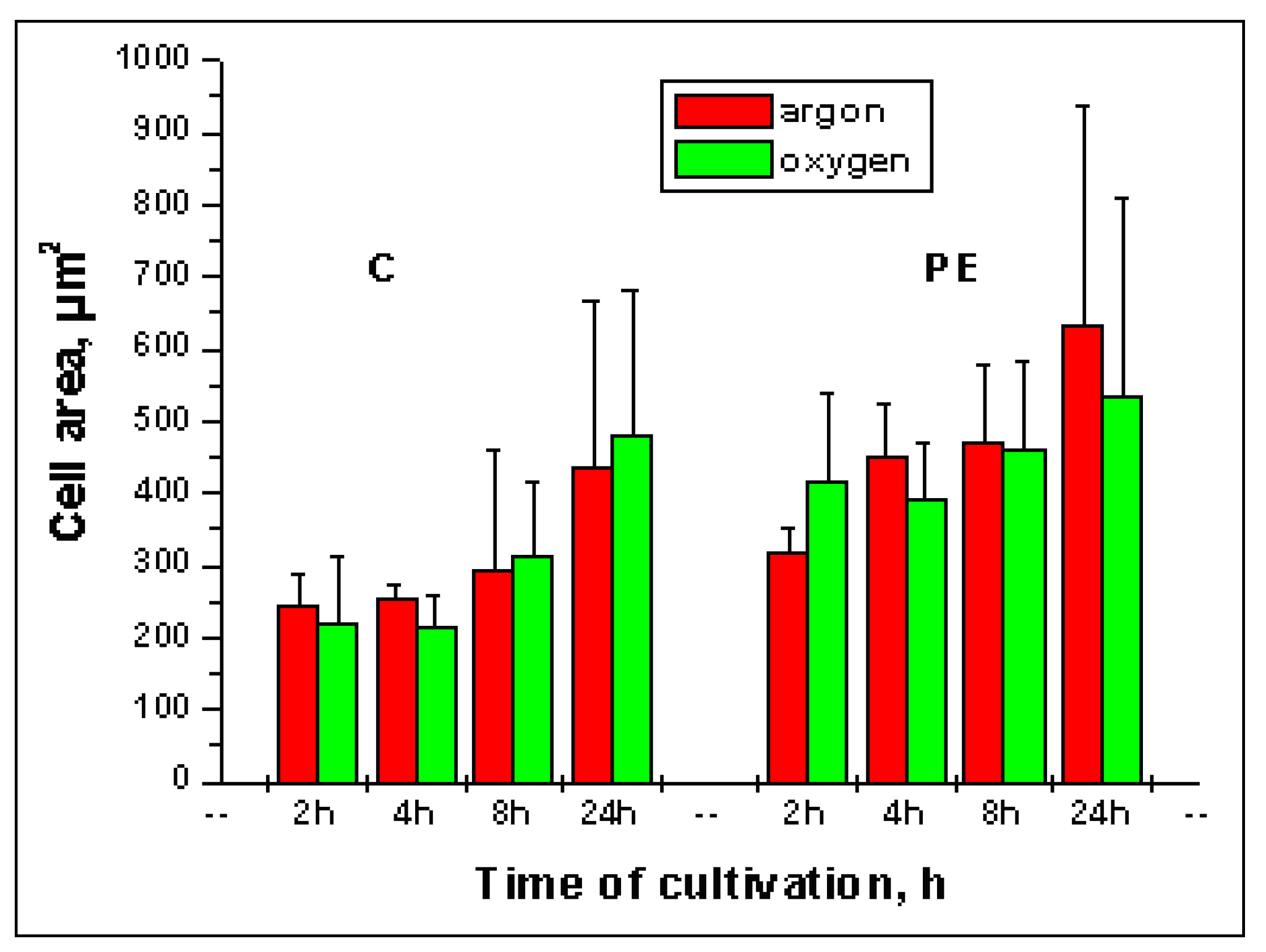
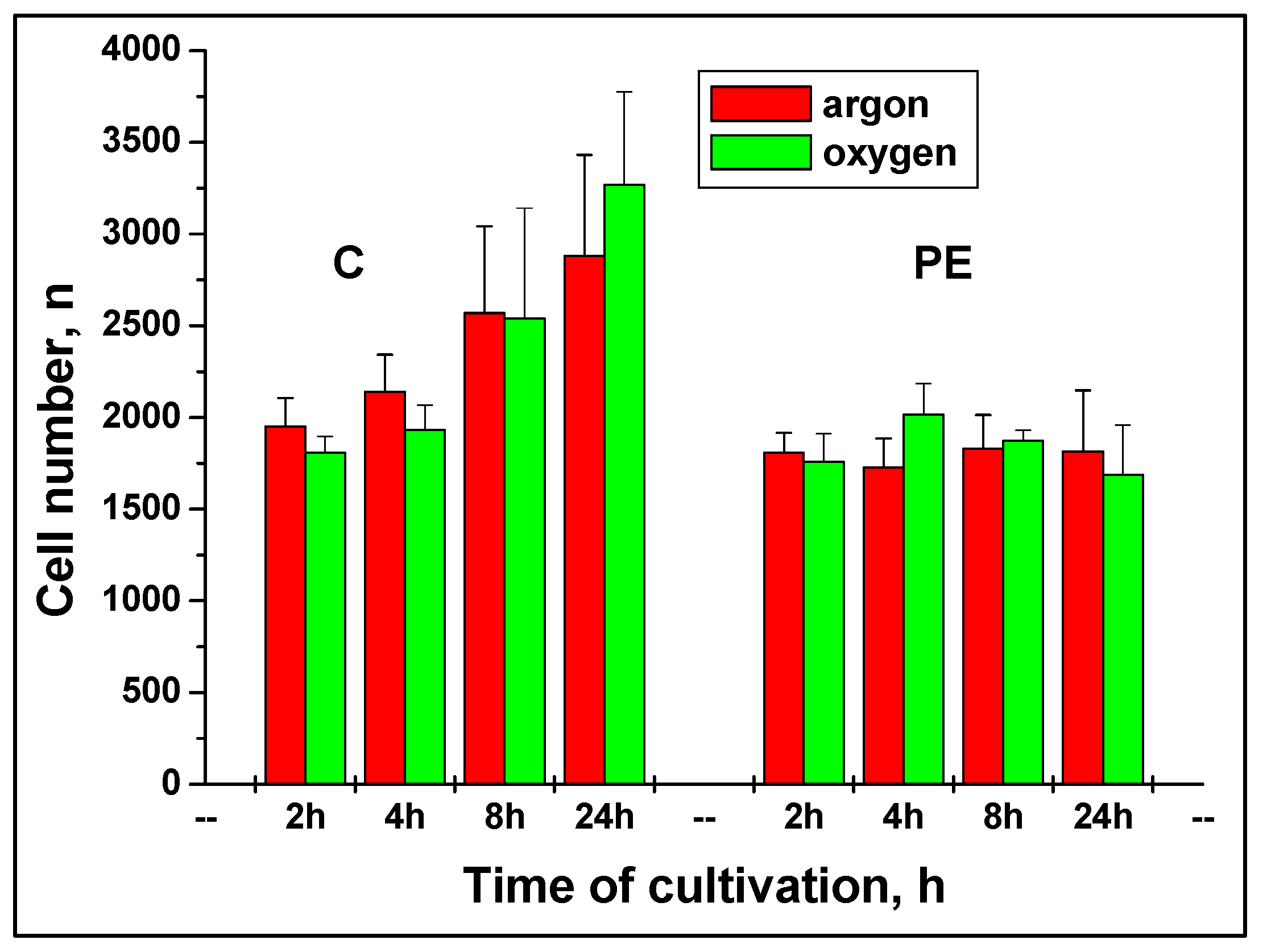
2.4. Advantages of developed method
2.4.1. Contrast optimization of BSE-images
2.4.2. Automated BSE-imaging analysis
3. Experimental Section
3.1. Substrate preparation
3.2. Cell culture
3.3. Cell viability/Membrane integrity test
3.4. Scanning electron microscopy (SEM)
3.5. Quantification of cell spreading
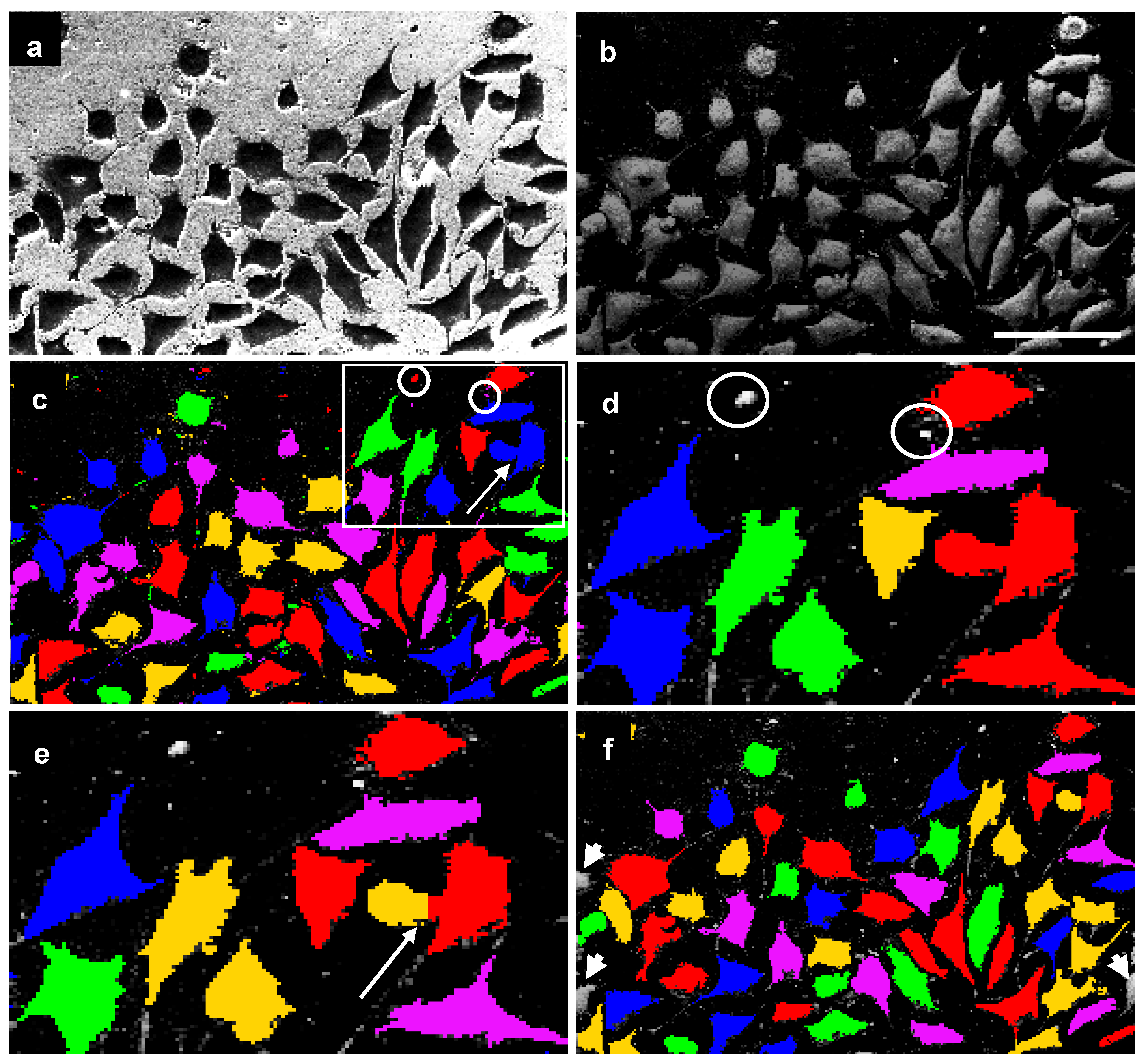
- Creation of an inverse image.
- Generation of background image information and its subtraction from the original image. In processing a non-uniform background, a smooth brightness gradient can be subtracted out. To generate an image with only this gradient, a mean filter with a large mask across a whole image was optimal [Figure 6(b)].
- Application of threshold segmentation. After this, discrete pixels were clustered to cell objects (marked by different colors in Figure 6). The cells were added to object lists for further processing steps. The magnitude of threshold segmentation was automatically adapted to the particular images.
- Deletion of non-cell objects. Small blemishes were automatically removed using a size filter [Figure 6(c and d), objects marked with circles].
- Splitting of tangent cells by distance. In some cases, threshold segmentation cannot separate several tangent cells [Figure 6(c and e), arrow]. An algorithm based on evaluation of the distance of each point in the cell interior to the nearest cell border was used to split those cells.
- Removal of fragmentary border cells. Border cells with area located predominantly outside the image (above half area) were checked and removed [Figure 6(f)].
- Calculation of several figures of merit like cell count, total area of cells, etc.
3.6. Statistical analysis
4. Conclusions
Acknowledgements
References and notes
- Owen, G.R.; Meredith, D.O.; ap Gwynn, I.; Richards, R.G. Focal adhesion quantification- a new assay of material biocompatibility? Review. Eur. Cell Mater. 2005, 9, 85–96. [Google Scholar] [PubMed]
- Zimmermann, H.; Katsen, A.D.; Ihmig, F.R.; Durst, C.H.P.; Shirley, S.G.; Fuhr, G.R. First steps of interdisciplinary approach towards miniaturised cryopreservation for cellular nanobiotechnology. IEEE Proc. Nanobiotechnol. 2004, 151, 134–138. [Google Scholar] [CrossRef]
- Curtis, A.S. The mechanism of adhesion of cells to glass. A study by interference reflection microscopy. J. Cell Biol. 1964, 20, 199–215. [Google Scholar] [CrossRef] [PubMed]
- Rajaraman, R.; Rounds, D.E.; Yen, S.P.S.; Rembaum, A. A scanning electron microscope study of cell adhesion and spreading in vitro. Exp. Cell Res. 1974, 88, 327–339. [Google Scholar] [CrossRef] [PubMed]
- Axelrod, D. Cell-substrate contacts illuminated by total internal reflection fluorescence. J. Cell Biol. 1981, 89, 141–145. [Google Scholar] [CrossRef] [PubMed]
- Rovensky, Yu.A.; Samoilov, V.I. Morphogenetic response of cultured normal and transformed fibroblasts, and epitheliocytes, to a cylindrical substratum surface. Possible role for the actin filament bundle pattern. J. Cell Sci. 1994, 107, 1255–1263. [Google Scholar] [PubMed]
- Meredith, D.O.; Owen, G.R.; ap Gwynn, I.; Richards, R.G. Variation in cell-substratum adhesion in relation to cell cycle phases. Exp. Cell Res. 2004, 293, 58–67. [Google Scholar] [CrossRef] [PubMed]
- Bigerelle, M.; Anselme, K. A kinetic approach to osteoblast adhesion on biomaterial surface. J. Biomed. Mater. Res. A 2005, 75, 530–540. [Google Scholar] [CrossRef] [PubMed]
- Curtis, A.S.; Wilkinson, C.D. Topographical control of cells. Biomaterials 1997, 18, 1573–1583. [Google Scholar]
- Curtis, A.S.; Wilkinson, C.D. Reactions of cells to topography. J. Biomater. Sci. Polymer Ed. 1998, 9, 1313–1329. [Google Scholar] [CrossRef]
- Chen, C.S.; Mrksich, M.; Huang, S.; Whitesides, G.M.; Ingber, D.E. Geometric control of cell life and death. Science 1997, 276, 1425–1428. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.S.; Mrksich, M.; Huang, S.; Whitesides, G.M.; Ingber, D.E. Micropatterned surfaces for control of cell shape, position and function. Biotechnol. Prog. 1998, 14, 356–363. [Google Scholar] [CrossRef] [PubMed]
- Dalby, M.J.; Riehle, M.O.; Johnstone, H.J.H.; Affrossman, S.; Curtis, A.S. Polymer-demixed nanotopography: Control of fibroblast spreading and proliferation. Tissue Eng. 2002, 8, 1099–1108. [Google Scholar] [CrossRef] [PubMed]
- Dalby, M.J.; Yarwood, S.J.; Riehle, M.O.; Johnstone, H.J.H.; Affrossman, S.; Curtis, A.S. Increasing fibroblast response to materials using nanotopography: Morphological and genetic measurements of cell response to 13-nm-high polymer demixed islands. Exp. Cell Res. 2002, 276, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Andersson, A.S.; Olsson, P.; Lidberg, U.; Sutherland, D. The effects of continuous and discontinuous groove edges on cell shape and alignment. Exp. Cell Res. 2003, 288, 177–188. [Google Scholar] [CrossRef] [PubMed]
- Rice, J.M.; Hunt, J.A.; Gallagher, J.A.; Hanarp, P.; Sutherland, D.S.; Gold, J. Quantitative assessment of the response of primary derived human osteoblasts and macrophages to a range of nanotopography surfaces in a single culture model in vitro. Biomaterials 2003, 24, 4799–4818. [Google Scholar] [CrossRef] [PubMed]
- Berry, C.C.; Campbell, G.; Spadiccino, A.; Robertson, M.; Curtis, A.S. The influence of microscale topography on fibroblasts attachment and motility. Biomaterials 2004, 25, 5781–5788. [Google Scholar] [CrossRef] [PubMed]
- Yim, E.K.; Reano, R.M.; Pang, S.W.; Yee, A.F.; Chen, C.F.; Leong, K.W. Nanopattern-induced changes in morphology and motility of smooth muscle cells. Biomaterials 2005, 26, 5405–5413. [Google Scholar] [CrossRef] [PubMed]
- Andersson, A.S.; Brink, J.; Lidberg, U.; Sutherland, D.S. Influence of systematically varied nanoscale topography on the morphology of epithelial cells. IEEE Trans. Nanobioscience 2003, 2, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Andersson, A.S.; Bäckhed, F.; von Euler, A.; Richter-Dahlfors, A.; Sutherland, D.; Kasemo, B. Nanoscale features influence epithelial cell morphology and cytokine production. Biomaterials 2003, 24, 3427–3436. [Google Scholar] [CrossRef] [PubMed]
- Dalby, M.J.; Gadegaard, N.; Riehle, M.O.; Wilkinson, C.D.W.; Curtis, A.S.G. Investigation filopodia sensing using arrays of defined nano-pits down to 35 nm diameter in size. Int. J. Biochem. Cell Biol. 2004, 36, 2005–2015. [Google Scholar] [CrossRef] [PubMed]
- Dalby, M.J.; Riehle, M.O.; Sutherland, D.S.; Agheli, H.; Curtis, A.S. Use of nanotopography to study mechano-transduction in fibroblasts – methods and perspectives. Eur. J. Cell Biol. 2004, 83, 159–169. [Google Scholar] [CrossRef] [PubMed]
- Dalby, M.J.; Riehle, M.O.; Sutherland, D.S.; Agheli, H.; Curtis, A.S. Morphological and microarray analysis of human fibroblasts cultured on nanocolumns produced by colloidal lithography. Eur. Cell Mater. 2005, 9, 1–8. [Google Scholar] [PubMed]
- Anselme, K.; Bigerelle, M. Modelling approach in cell/meterial interactions studies. Biomaterials 2006, 27, 1187–1199. [Google Scholar] [CrossRef] [PubMed]
- Curtis, A.S. Cell reactions with biomaterials: the microscopies. Eur. Cell Mater. 2001, 1, 59–65. [Google Scholar] [PubMed]
- Richards, R.G.; Owen, G.R.; Rahn, B.A.; ap Gwynn, I. A quantitative method of measuring cell adhesion areas. Cells Mater. 1997, 7, 15–30. [Google Scholar]
- Baxter, L.; Frauchiger, V.; Textor, M.; ap Gwynn, I.; Richards, R.G. Fibroblast and osteoblast adhesion and morphology on calcium phosphate surfaces. Eur. Cell Mater. 2002, 4, 1–17. [Google Scholar]
- Harris, L.G.; Patterson, L.M.; Bacon, C.; ap Gwynn, I.; Richards, R.G. Assessment of the cytocompatibility of different coated titanium surfaces to fibroblasts and osteoblasts. J. Biomed. Mater. Res. A 2005, 73, 12–20. [Google Scholar] [CrossRef]
- Richards, R.G.; ap Gwynn, I. Backscattered electron imaging of undersurface of resin-embedded cells by field emission scanning electron microscopy. J. Microsc. 1995, 177, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Harris, L.G.; ap Gwynn, I.; Richards, R.G. Contrast optimisation for backscattered electron imaging of resin embedded cells. Scanning Microscopy 1999, 13, 71–81. [Google Scholar]
- Park, J-W.; Kim, Y-J.; Park, C.H.; Lee, D-H.; Ko, Y.G.; Jang, J-H.; Lee, C.S. Enhanced osteoblast response to an equal channel angular pressing-processed pure titanium substrate with microrough surface topography. Acta Biomaterial. 2009. [Google Scholar] [CrossRef]
- Sammons, R.L.; Lumbikanonda, N.; Gross, M.; Cantzler, P. Comparasion of osteoblast spreading on microstructured dental implant surfaces and cell behaviour in an explant model of osseointegration: A scanning electron microscopic study. Clin. Oral Implan. Res. 2005, 16, 657–666. [Google Scholar] [CrossRef]
- Hallab, N.J.; Bundy, K.J.; O`Connor, K.; Moses, R.L.; Jacobs, J.J. Evaluation of metallic and polymeric biomaterial surface energy and surface roughness characteristics for directed cell adhesion. Tissue Eng. 2001, 7, 55–71. [Google Scholar] [CrossRef] [PubMed]
- Merrett, K.; Cornelius, R.M.; McClung, W.G.; Unsworth, L.D.; Sheardown, H. Surface analysis methods for characterizing polymeric biomaterials. J. Biomater. Sci. Polym. Ed. 2002, 13, 593–622. [Google Scholar] [CrossRef] [PubMed]
- Chu, P.K.; Chen, J.Y.; Wang, L.P.; Huang, N. Plasma surface modification of biomaterials. Mater. Sci. Eng. 2002, R36, 143–206. [Google Scholar] [CrossRef]
- Ŝvorĉík, V.; Kaśalková, N.; Slepička, P.; Záruba, K.; Král, V.; Bačáková, L.; Pařízek, M.; Lisá, V.; Ruml, T.; Gbelcová, H.; Rimpelová, S.; Macková, A. Cytocompatibility of Ar+ plasma treated and Au nanoparticle-grafted PE. Nucl. Instrum. Meth. B 2009, 267, 1904–1910. [Google Scholar] [CrossRef]
- Zimmermann, H.; Richter, E.; Reichle, C.; Westpal, I.; Geggier, P.; Rehn, U.; Rogaschewski, S.; Bleiß, W.; Fuhr, G.R. Mammalian cell traces- morphology, molecular composition, artificial guidance and biotechnological relevance as a new type of “bionanotube”. Appl. Phys. A 2001, 73, 11–26. [Google Scholar] [CrossRef]
- Hallab, N.J.; Bundy, K.J.; O`Connor, K.; Clark, R.; Moses, R.L. Surface charge, biofilm composition and cellular morphology as related to cellular adhesion to biomaterials. In Proceedings 14th Southern Biomedical Engineering Conference, Shreveport, LA, USA, April 7-9; 1995; pp. 81–84. [Google Scholar]
- Gallagher, J.O.; McGhee, K.F.; Wilkinson, C.D.; Riehle, M.O. Interaction of animal cells with ordered nanotopography. IEEE Trans. Nanobioscience 2002, 1, 24–28. [Google Scholar] [CrossRef] [PubMed]
- Boucrot, E.; Kirchhausen, T. Endosomal recycling controls plasma membrane area during mitosis. Proc. Natl. Acad. Sci. USA 2007, 104, 7939–7944. [Google Scholar] [CrossRef]
- Katsen, A.D.; Tregubova, N.A.; Andrushkevich, V.V.; Rovensky, Yu.A. Identification and study of the same cell in monolayer culture by different methods of light microscopy, scanning and transmission electron microscopy. Application for cryodamaged cells. Scanning 1992, 14, 225–230. [Google Scholar] [CrossRef]
- Katsen, A.D.; Vollmar, B.; Mestres-Ventura, P.; Menger, M.D. Cell surface and nuclear changes during TNF-α-induced apoptosis in WEHI 164 murine fibrosarcoma cells. Virchows Arch 1998, 433, 75–83. [Google Scholar] [CrossRef] [PubMed]
- Dankberg, F.; Persidsky, M.D. A test of granulocyte membrane integrity and phagocytic function. Cryobiology 1976, 13, 430–432. [Google Scholar] [CrossRef] [PubMed]
© 2009 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Katsen-Globa, A.; Peter, L.; Zöllner, S.; Dörge, T.; Daffertshofer, M.; Preckel, H.; Schmitt, D.; Zimmermann, H. A Novel Approach for Automated Analysis of Cell Attachment and Spreading Based on Backscattered Electron Imaging by Scanning Electron Microscopy. Materials 2009, 2, 1402-1416. https://doi.org/10.3390/ma2031402
Katsen-Globa A, Peter L, Zöllner S, Dörge T, Daffertshofer M, Preckel H, Schmitt D, Zimmermann H. A Novel Approach for Automated Analysis of Cell Attachment and Spreading Based on Backscattered Electron Imaging by Scanning Electron Microscopy. Materials. 2009; 2(3):1402-1416. https://doi.org/10.3390/ma2031402
Chicago/Turabian StyleKatsen-Globa, Alisa, Leonora Peter, Susan Zöllner, Thomas Dörge, Martin Daffertshofer, Hartwig Preckel, Daniel Schmitt, and Heiko Zimmermann. 2009. "A Novel Approach for Automated Analysis of Cell Attachment and Spreading Based on Backscattered Electron Imaging by Scanning Electron Microscopy" Materials 2, no. 3: 1402-1416. https://doi.org/10.3390/ma2031402



