Application of Nanodiamonds in Biomolecular Mass Spectrometry
Abstract
:1. Introduction
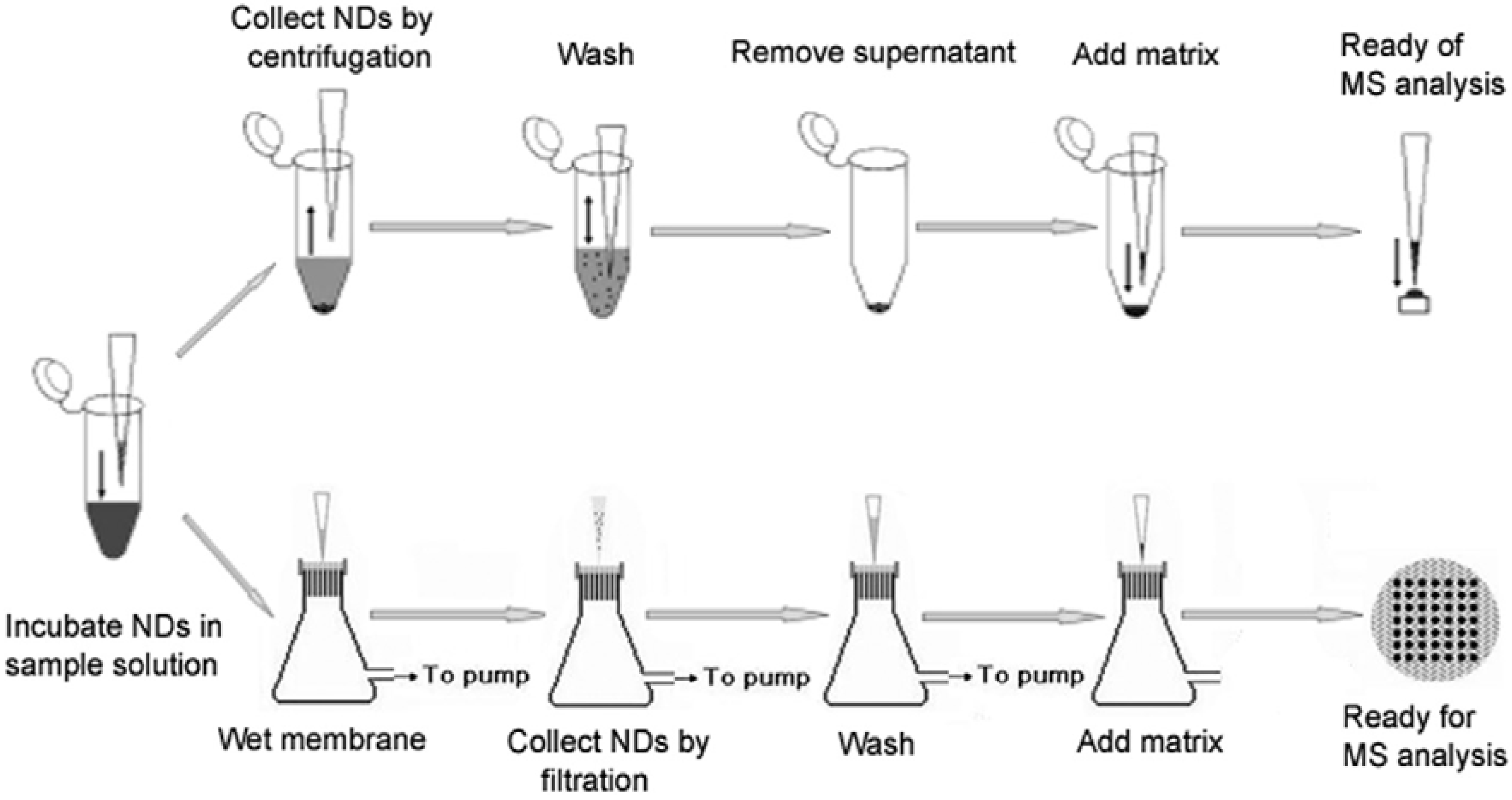
2. Methods
3. Applications of Carboxylated/Oxidized NDs
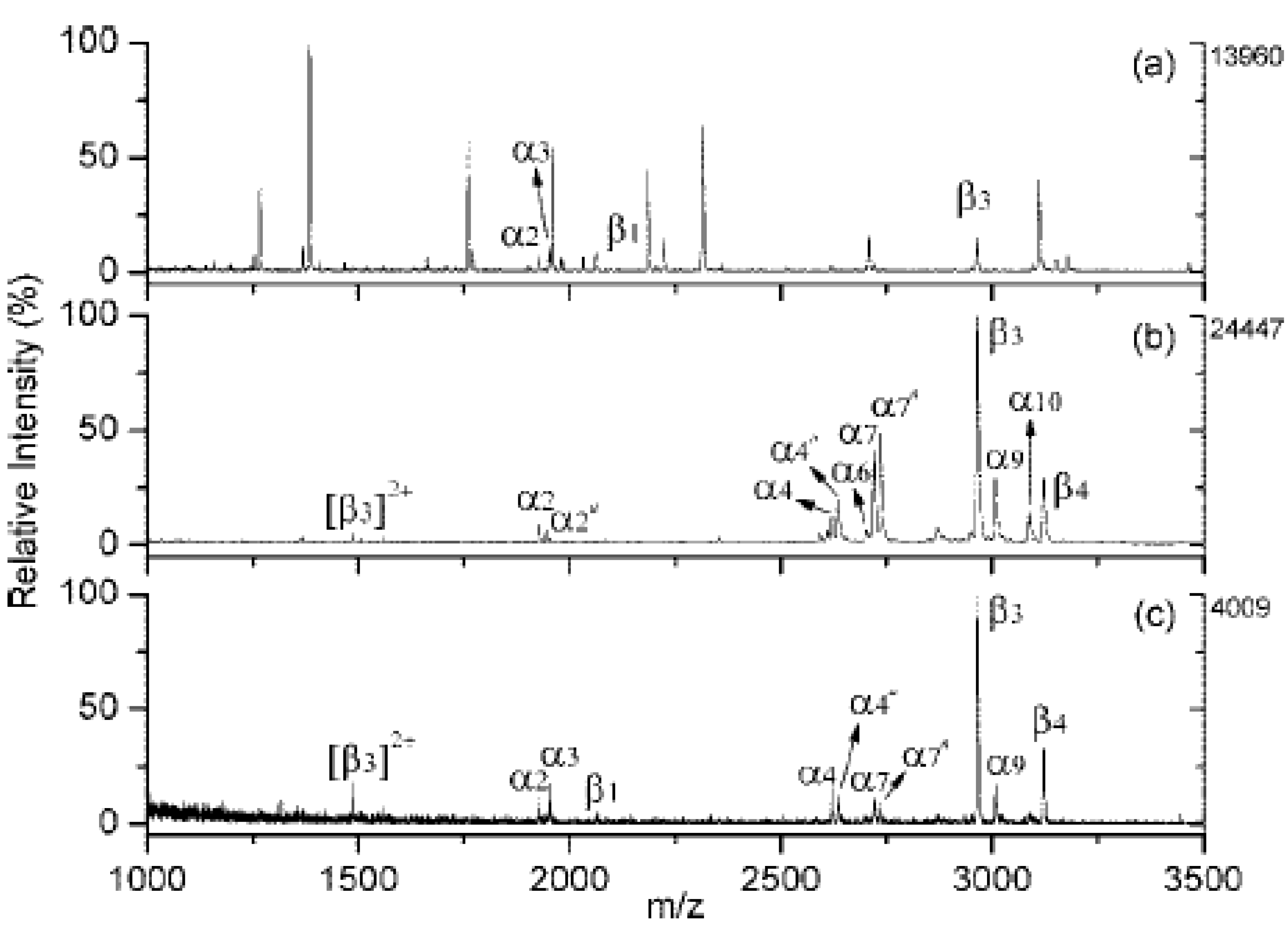
3.1. Analysis of Peptides
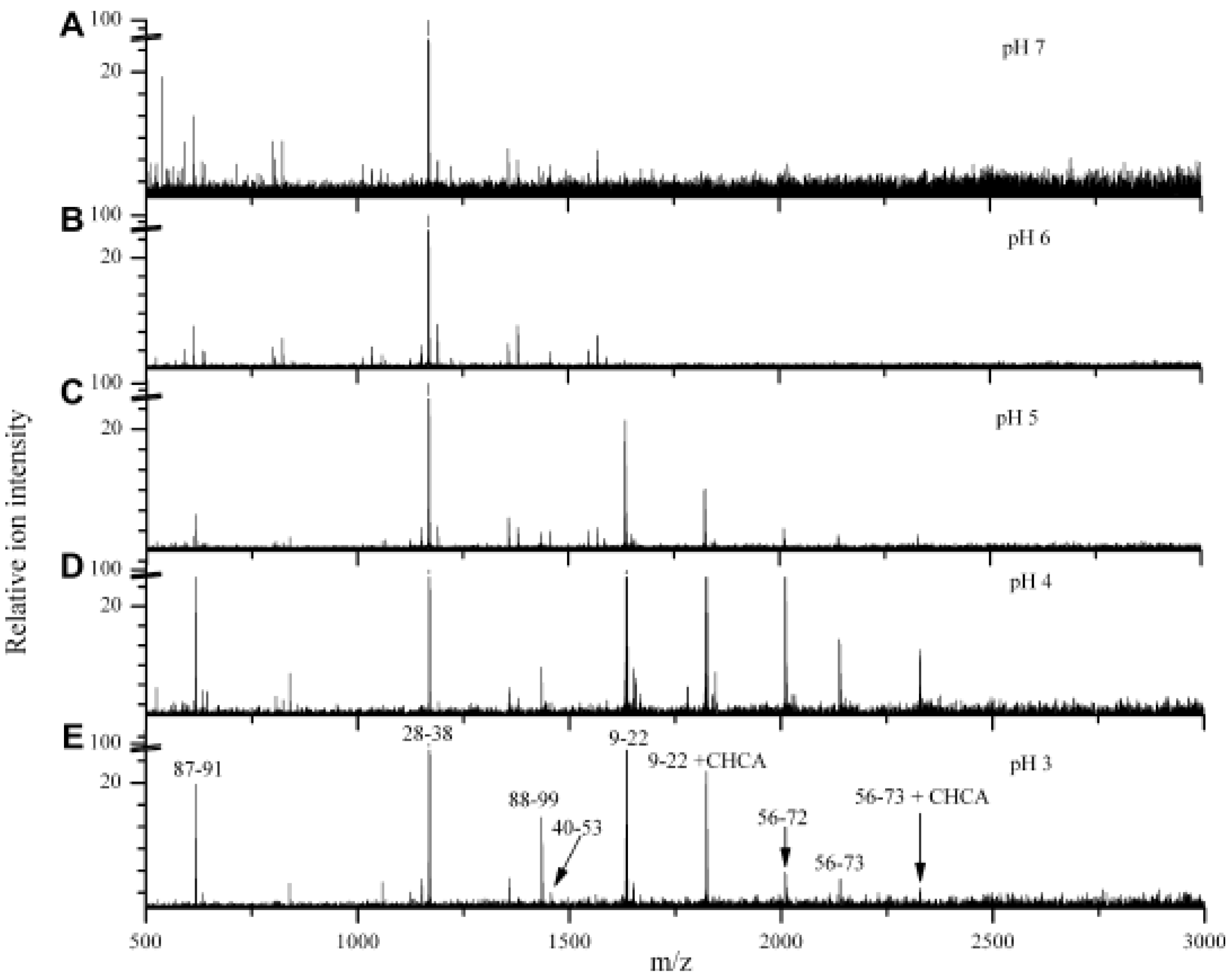
3.2. Analysis of Proteins

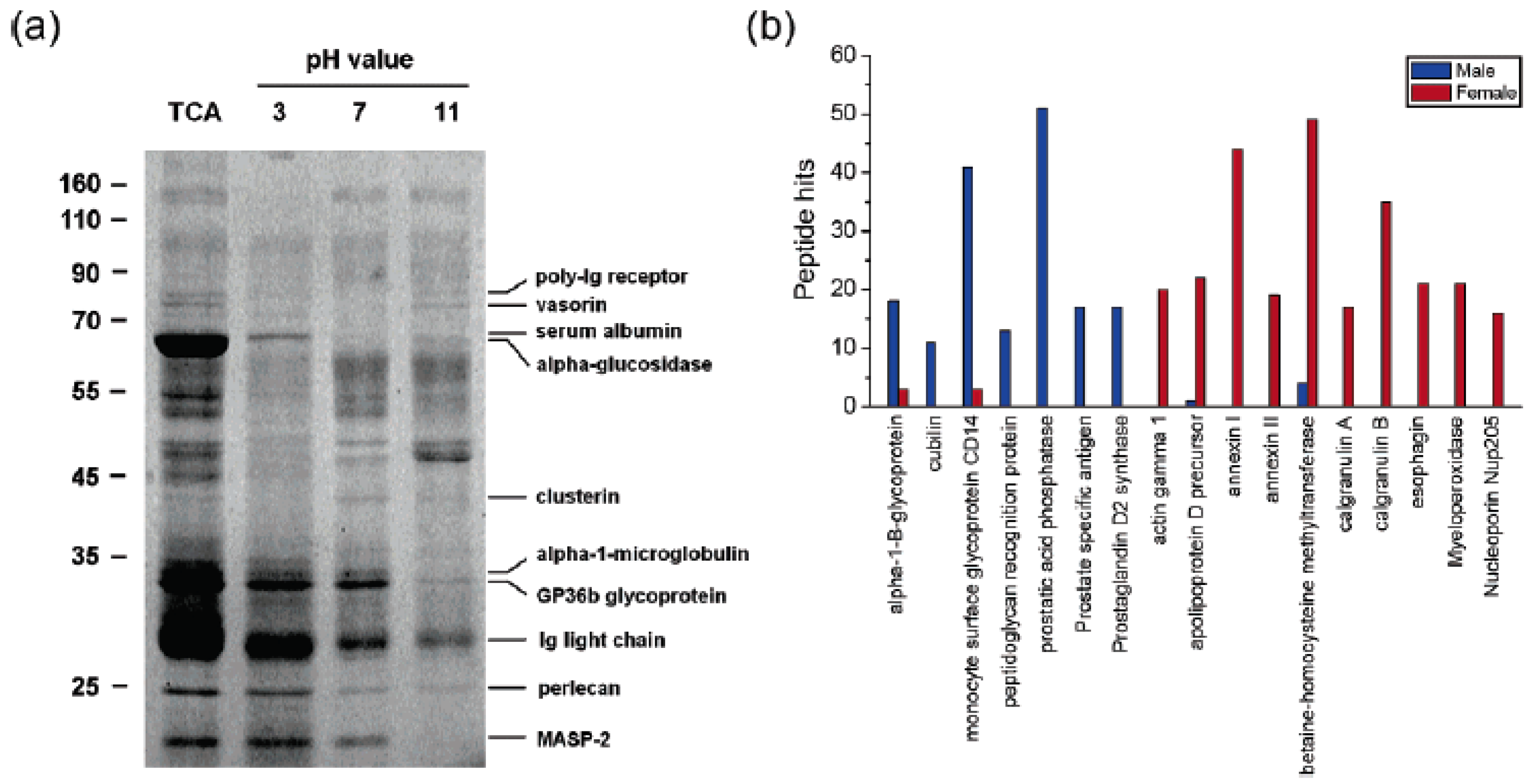
3.3. Analysis of Body Fluids
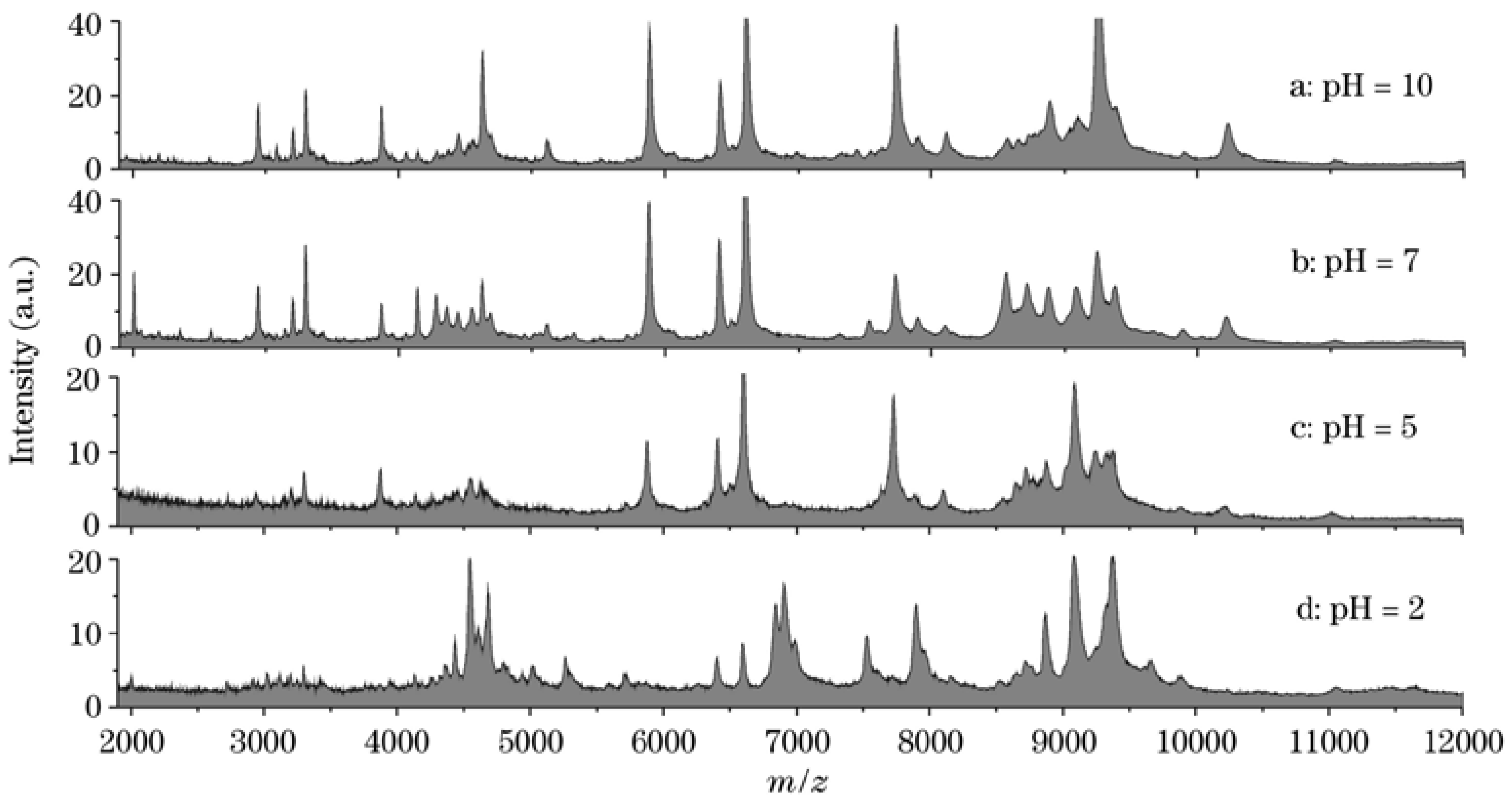
3.4. Other Applications

4. Applications of Surface-Modified NDs
4.1. Polylysine (PL)-Coated NDs
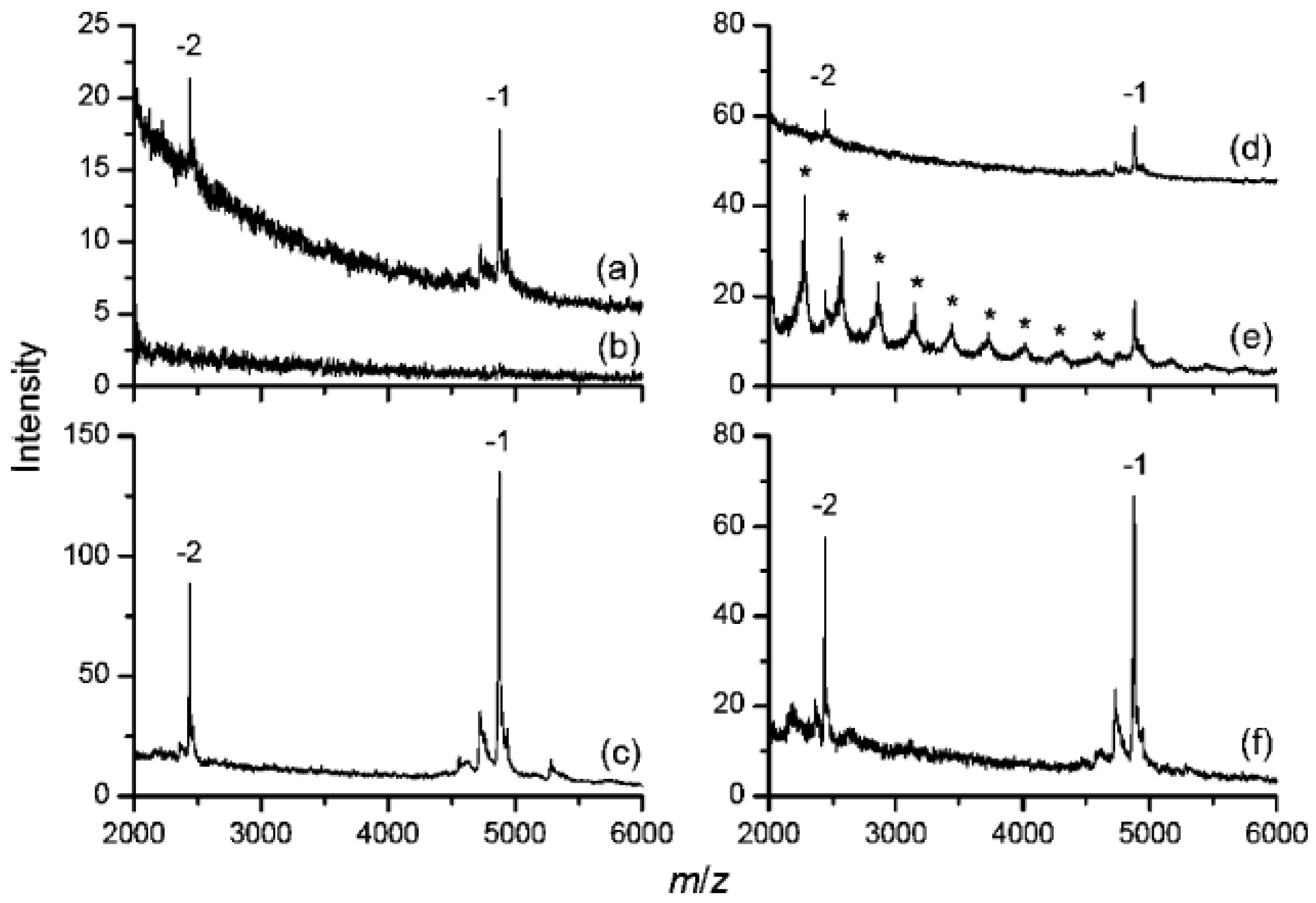
4.2. Polyarginine (PA)-Coated NDs
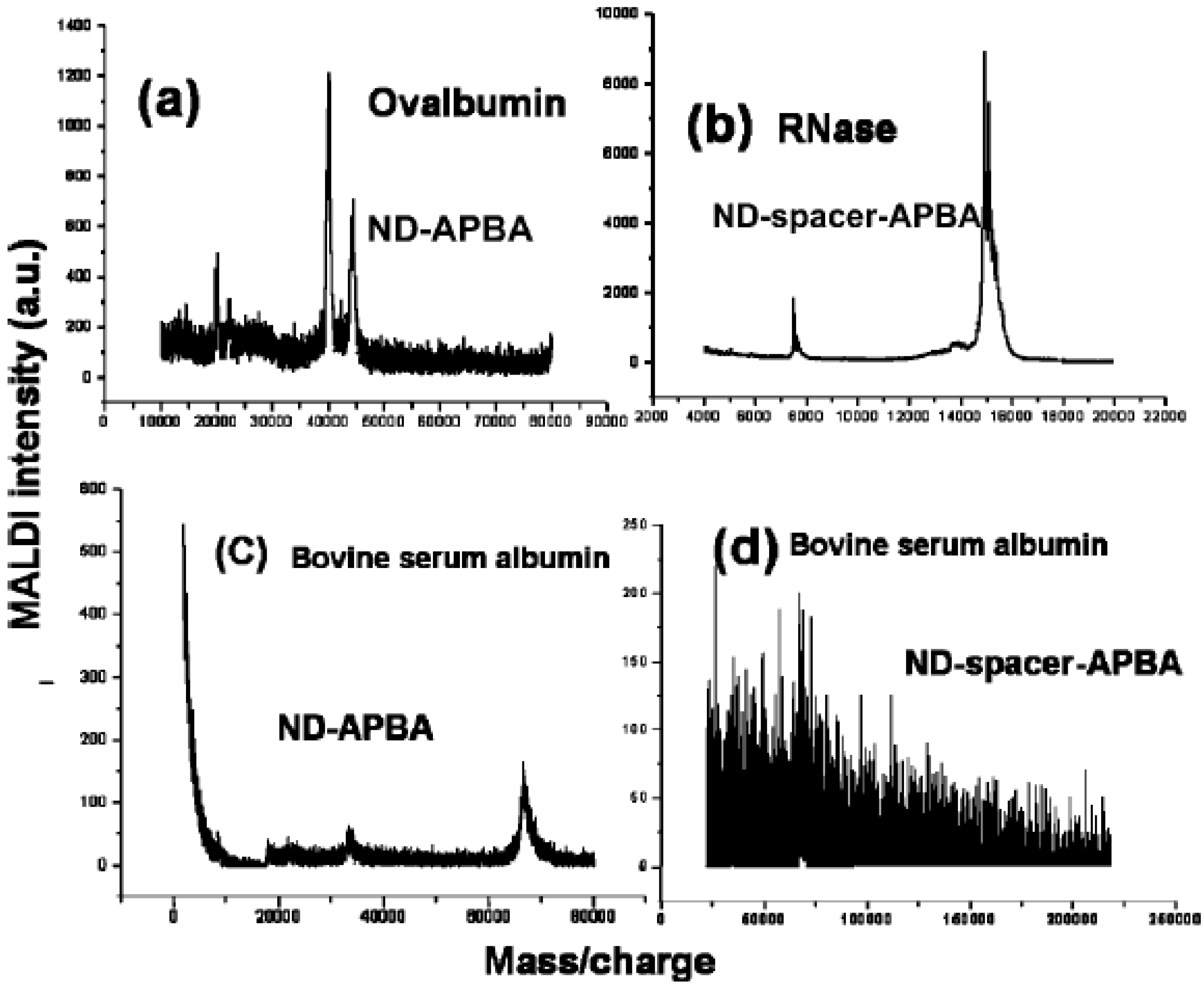
4.3. Modifications by Aminophenylboronic Acid (APBA)
5. NDs as Platforms in MS-Based Study
5.1. Digestion on the Surfaces of NDs
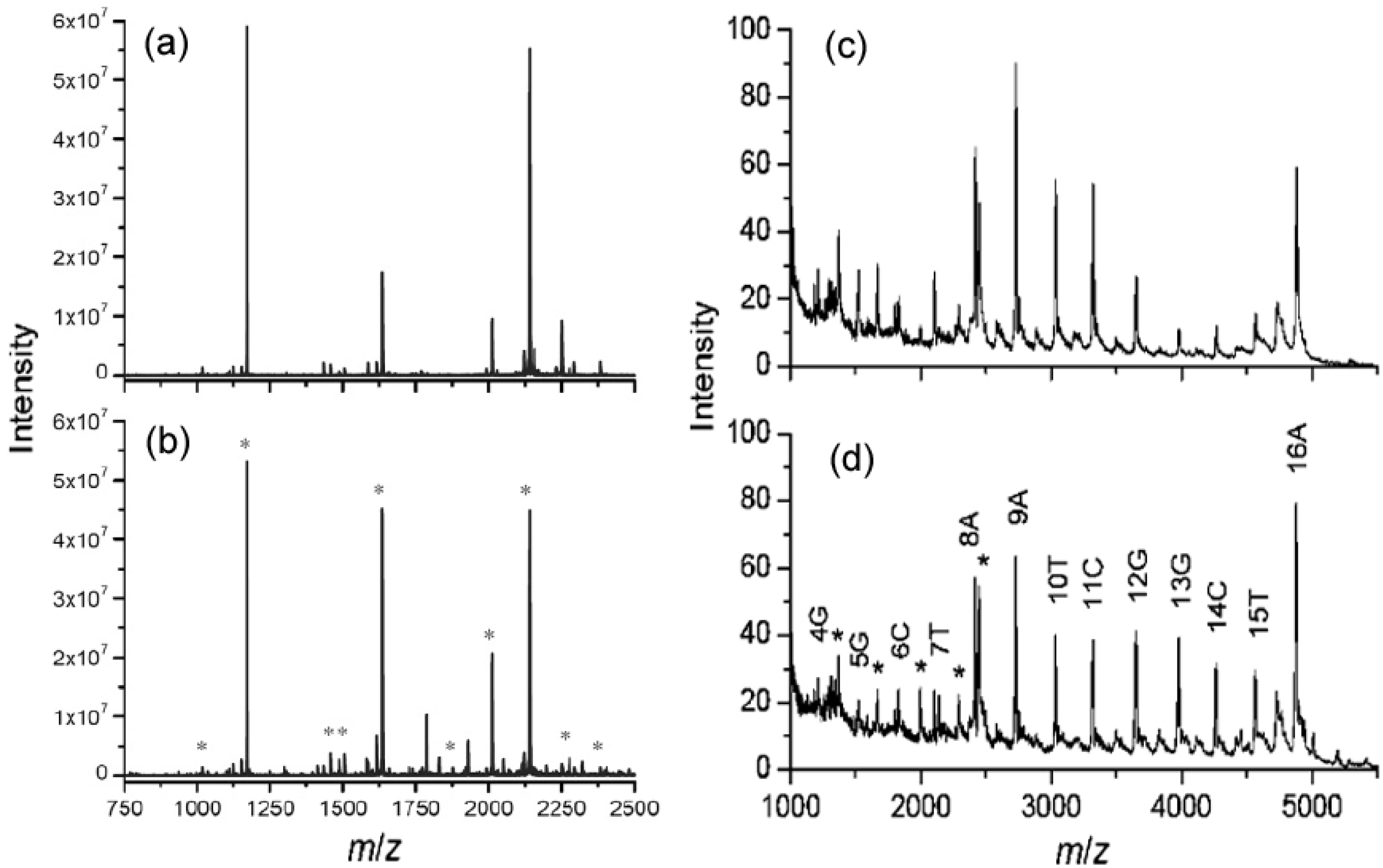
5.2. SPEED Method
5.3. Non-Covalent Interaction Study
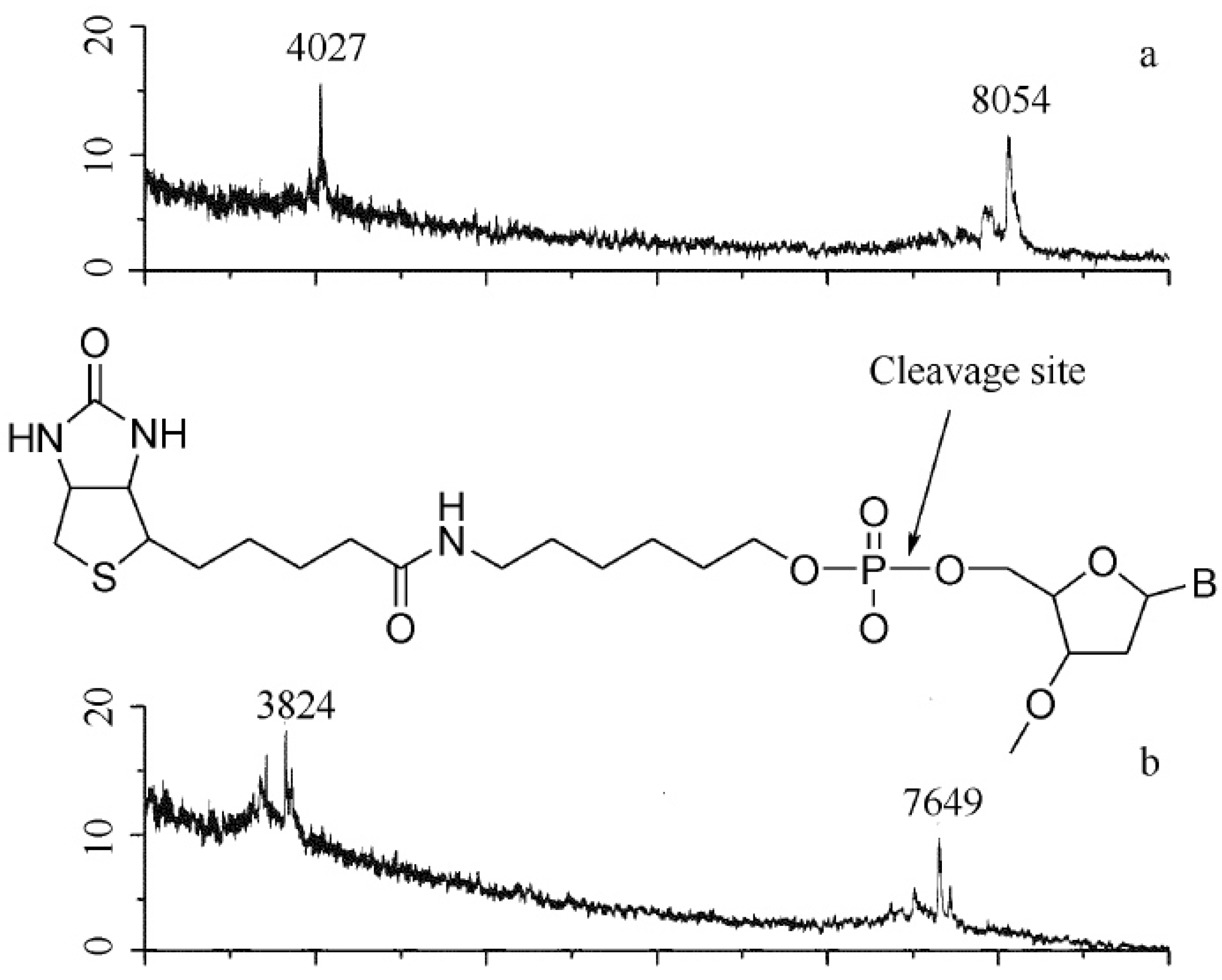
6. Other MS Study
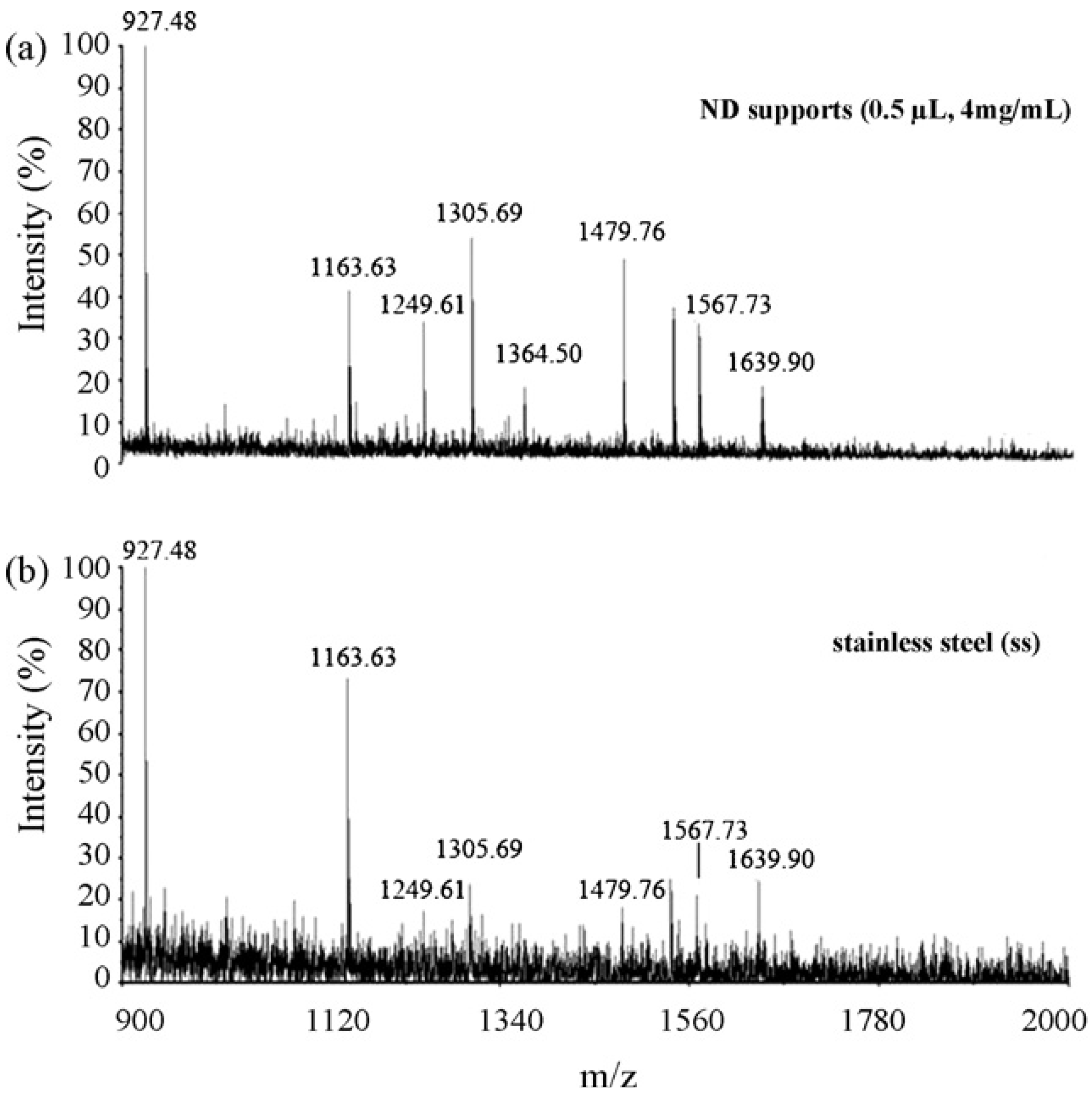
7. What is the Next?
8. Conclusions
Acknowledgements
References and Notes
- Danilenko, V.V. On the history of the discovery of nanodiamond synthesis. Phys. Sol. State 2004, 46, 595–599. [Google Scholar] [CrossRef]
- Lewis, R.S.; Ming, T.; Wacker, J.F.; Anders, E.; Steel, E. Interstellar diamonds in meteorites. Nature 1987, 326, 160–162. [Google Scholar] [CrossRef]
- Holt, K.B. Diamond at the nanoscale: Applications of diamond nanoparticles from cellular biomarkers to quantum computing. Phil. Trans. R. Soc. A 2007, 365, 2845–281. [Google Scholar] [CrossRef] [PubMed]
- Krueger, A. Diamond nanoparticles: Jewels for chemistry and physics. Adv. Mater. 2008, 20, 2445–2449. [Google Scholar] [CrossRef]
- Barnard, A.S. Diamond standard in diagnostics: Nanodiamond biolabels make their mark. Analyst 2009, 134, 1751–1764. [Google Scholar] [CrossRef] [PubMed]
- Baidakova, M.; Vul, A. New prospects and frontiers of nanodiamond clusters. J. Phys. D: Appl. Phys. 2007, 40, 6300–6311. [Google Scholar] [CrossRef]
- Gruber, A.; Drabenstedt, A.; Tietz, C.; Fleury, L.; Wrachtrup, J.; von Borczyskowski, C. Scanning confocal optical microscopy and magnetic resonance on single defect centers. Science 1997, 276, 2012–2014. [Google Scholar] [CrossRef]
- Yu, S.J.; Kang, M.W.; Chang, H.C.; Chen, K.M.; Yu, Y.C. Bright fluorescent nanodiamonds: No photobleaching and low cytotoxicity. J. Am. Chem. Soc. 2005, 127, 17604–17605. [Google Scholar] [CrossRef] [PubMed]
- Fu, C.C.; Lee, H.Y.; Chen, K.; Lim, T.S.; Wu, H.Y.; Lin, P.K.; Wei, P.K.; Tsao, P.H.; Chang, H.C.; Fann, W. Characterization and application of single fluorescent nanodiamonds as cellular biomarkers. Proc. Natl. Acad. Sci. USA 2007, 104, 727–732. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.K.; Chen, F.; Chen, P.Y.; Lee, T.J.F.; Cheng, C.L.; Chang, C.C.; Ho, Y.P.; Chao, J.I. Alpha-bungarotoxin binding to target cell in a developing visual system by carboxylated nanodiamond. Nanotechnology 2008, 19, 205102. [Google Scholar] [CrossRef] [PubMed]
- Karas, M.; Hillenkamp, F. Laser desorption ionization of proteins with molecular masses exceeding 10000 daltons. Anal. Chem. 1988, 60, 2299–2301. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, K.; Waki, H.; Ido, Y.; Akita, S.; Yoshida, Y.; Yoshida, T.; Matsuo, T. Protein and polymer analyses up to m/z 100 000 by laser ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrum 1988, 2, 151–153. [Google Scholar] [CrossRef]
- Andersen, J.S.; Svensson, B.; Roepstorff, P. Electrospray ionization and matrix assisted laser desorption/ionization mass spectrometry: Powerful analytical tools in recombinant protein chemistry. Nat. Biotechnol. 1996, 14, 449–457. [Google Scholar] [CrossRef] [PubMed]
- Hillenkamp, F.; Karas, M.; Beavis, R.C.; Chait, B.T. Matrix-assisted laser desorption/ionization mass spectrometry of biopolymers. Anal. Chem. 1991, 63, 1193A–1203A. [Google Scholar] [CrossRef] [PubMed]
- Warren, M.E.; Brockman, A.H.; Ron Orlando, R. On-probe solid-phase extraction/MALDI-MS using ion-pairing interactions for the cleanup of peptides and proteins. Anal. Chem. 1998, 70, 3757–3761. [Google Scholar]
- Xu, Y.; Bruening, M.L.; Watson, J.T. Non-specific, on-probe cleanup methods for MALDI-MS samples. Mass Spectrum. Rev. 2003, 6, 429–440. [Google Scholar] [CrossRef]
- Kong, X.; Sabu, S. Rapid MALDI mass spectrometric analysis with prestructured membrane filters and functionalized diamond nanocrystlas. Anal. Chim. Acta. 2009. [Google Scholar] [CrossRef]
- Fenn, J.B.; Mann, M.; Meng, C.K.; Wong, S.F.; Whitehouse, C.M. Electrospray ionization for mass spectrometry of large biomolecules. Science 1989, 246, 64–71. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.H.; Lee, S.C.; Sabu, S.; Fang, H.C.; Chung, S.C.; Han, C.C.; Chang, H.C. Solid-phase extraction and elution on diamond(SPEED): A fast and general platform for proteome analysis with mass spectrometry. Anal. Chem. 2006, 78, 4228–4234. [Google Scholar] [CrossRef] [PubMed]
- Kong, X.L.; Huang, L.C.L.; Hsu, C.M.; Han, C.C.; Chang, H.C. High-affinity capture of proteins by diamond nanoparticles for mass spectrometric analysis. Anal. Chem. 2005, 77, 259–265. [Google Scholar] [CrossRef] [PubMed]
- Kong, X.L. Application of nanodiamonds in human body fluid analysis by matrix-assisted laser desorption/ionization mass spectrometry. Chin. Opt. Lett. 2008, 6, 417–420. [Google Scholar] [CrossRef]
- Ushizawa, K.; Sato, Y.; Mitsumori, T.; Machinami, T.; Ueda, T.; Ando, T. Covalent immobilization of DNA on diamond and its verification by diffuse reflectance infrared spectroscopy. Chem. Phys. Lett. 2002, 351, 105–108. [Google Scholar] [CrossRef]
- Huang, L.C.L.; Chang, H.C. Adsorption and immobilization of cytochrome C on nanodiamonds. Langmuir 2004, 20, 5879–5884. [Google Scholar] [CrossRef] [PubMed]
- Sabu, S.; Yang, F.C.; Wang, Y.S; Chen, W.H; Chou, M.I.; Chang, H.C.; Han, H.C. Peptide analysis: Solid phase extraction-elution on diamond combined with atmospheric pressure matrix-assisted laser desorption/ionization-Fourier transform ion cyclotron resonance mass spectrometry. Anal. Biochem. 2007, 367, 190–200. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.T.B.; Chang, H.C.; Wu, V.W.K. Adsorption and hydrolytic activity of lysozyme on diamond nanocrystallites. Diamond Relat. Mater. 2007, 16, 872–876. [Google Scholar] [CrossRef]
- Bondar, V.S.; Pozdnyakova, I.O.; Puzyr, A.P. Applications of nanodiamonds for the separation and purification of proteins. Phys. Sol. State. 2004, 46, 758–760. [Google Scholar] [CrossRef]
- Hutchens, T.W.; Yip, T.T. New desorption strategies for the mass spectrometric analysis of macromolecules. Rapid Commun. Mass Spectrom. 1993, 7, 576–580. [Google Scholar] [CrossRef]
- Petricoin, E.F.; Ardekani, A.M.; Hitt, V.A.; Levine, P.J.; fusaro, V.A.; Steinberg, S.M.; Mills, G.B.; Simone, C.; Fishman, D.A.; Kohn, E.C.; Liotta, L.A. Use of proteomic patterns in serum to identify ovarian cancer. Lancet 2002, 359, 572–577. [Google Scholar] [CrossRef] [PubMed]
- Najam-ul-Haq, M.; Rainer, M.; Szabo, Z.; Vallant, R.; Huch, C.W.; Bonn, G.K. Role of carbon nano-materials in the analysis of biological materials by laser desorption/ionization-mass spectrometry. J. Biochem. Biophys. Methods 2007, 70, 319–328. [Google Scholar] [CrossRef] [PubMed]
- Pluskal, M.G. Microscale sample preparation. Nat. Biotechnol. 2000, 18, 104–105. [Google Scholar] [CrossRef] [PubMed]
- Tzeng, Y.K.; Chang, C.C.; Huang, C.N.; Wu, C.C.; Han, C.C.; Chang, H.C. Facile MALDI-MS analysis of neutral glycans in NaOH-doped matrixes: Microwave-assisted deglycosylation and one-step purification with diamond nanoparticles. Anal. Chem. 2008, 80, 6809–6814. [Google Scholar]
- Kong, X.; Huang, L.C.L.; Liau, S.C.V.; Han, C.C.; Chang, H.C. Polylysine-coated diamond nanocrystals for MALDI-TOF mass analysis of DNA oligonucleotides. Anal. Chem. 2005, 77, 4273–4277. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.K.; Wu, C.C.; Wang, Y.S.; Chang, H.C. Selective extraction and enrichment of multiphosphorylated peptides using polyarginie-coated diamond nanoparticles. Anal. Chem. 2008, 80, 3791–3797. [Google Scholar] [CrossRef] [PubMed]
- Yeap, W.S.; Tan, Y.Y.; Loh, K.P. Using detonation nanodiamonds for the specific capture of glycoproteins. Anal. Chem. 2008, 80, 4659–4665. [Google Scholar] [CrossRef] [PubMed]
- Kong, X.L. Nanodiamonds used as a platform for studying noncovalent interaction by MALDI-MS. Chin. J. Chem. 2008, 26, 1811–1815. [Google Scholar] [CrossRef]
- Wei, L.M.; Shen, Q.; Lu, H.J.; Yang, P.Y. Pretreatment of low-abundance peptides on detonation nanodiamond for direct analysis by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. J. Chromatogr. B 2009, 877, 3631–3637. [Google Scholar] [CrossRef]
- Wei, L.M.; Xue, Y.; Zhou, X.W.; Jin, H.; Shi, Q.; Lu, H.J.; Yang, P.Y. Nanodiamond MALDI support for enhancing the credibility of identifying proteins. Talanta 2008, 74, 1363–1370. [Google Scholar] [CrossRef] [PubMed]
- Najam-ul-Haq, M.; Rainer, M.; Huch, C.W.; Hausberger, P.; kraushaar, H.; Bonn, G.K. Nanostructured diamond-like carbon on digital versatile disc as a matrix-free target for laser desorption/ionization mass spectrometry. Anal. Chem. 2008, 80, 7467–7472. [Google Scholar] [CrossRef] [PubMed]
- Blechova, P.; Havlova, P.; Gajdosova, D.; Havel, J. New possibilities of matrix-assisted laser desorption ionization time of flight mass spectrometry to analyze barley malt quality: Highly sensitive detection of mycotoxins. Environ. Toxicol. 2006, 21, 403–408. [Google Scholar] [CrossRef] [PubMed]
- Elosta, S.; Gajdosova, D.; Hegrova, B.; Havel, J. MALDI TOF mass spectrometry of selected mycotoxins in barley. J. Appl. Biomed. 2007, 5, 39–47. [Google Scholar]
- Houska, J.; Panyala, N.R.; Pena-Mendez, E.M.; Havel, J. Mass spectrometry of nanodiamonds. Rapid Commun. Mass Spectrom. 2009, 23, 1125–1131. [Google Scholar] [CrossRef] [PubMed]
- Shenderova, O.A.; Zhirnov, V.V.; Brenner, D.W. Carbon nanostructures. Crit. Rev. Solid State Mater. Sci. 2002, 27, 227–356. [Google Scholar] [CrossRef]
- Dolmatov, V.Y. Detonation synthesis ultradispersed diamonds: Properties and applications. Russ. Chem. Rev. 2001, 70, 607–626. [Google Scholar] [CrossRef]
- Krueger, A.; Ozawa, M.; Kataoka, F.; Fujino, T.; Suzuki, A.E.; Aleksenskii, A.E.; Vul, A.Y.; Osawa, E. Unusually tight aggregation in detonation nanodiamond: Identification and disintegration. Carbon 2005, 43, 1722–1730. [Google Scholar] [CrossRef]
- Ji, S.F.; Jiang, T.L.; Xu, K.; Li, S.B. FTIR study of the adsorption of water on ultradispersed diamond powder surface. Appl. Surf. Sci. 1998, 133, 231–238. [Google Scholar] [CrossRef]
- Zheng, W.W.; Hsieh, Y.H.; Chiu, Y.C.; Cai, S.J.; Cheng, C.L.; Cheng, C. Organic functionalization of ultradispersed nanodiamond: Synthesis and applications. J. Mater. Chem. 2009, 19, 8432–8441. [Google Scholar] [CrossRef]
- Hartl, A.; Schmich, E.; Garrido, J.A.; Hernando, J.; Catharino, S.C.R.; Walter, S.; Feulner, P.; Kromka, A.; Steinmuller, D.; Stutzmann, A. Protein-modified nanocrystalline diamond thin films for biosensor applications. Nat. Mater. 2004, 3, 736–742. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.J.; Song, K.S.; Nakamura, Y.; Ueno, T.; Funatsu, T.; Ohdomari, I.; Kawarada, H. DNA micropatterning on polycrystalline diamond via one-Step direct amination. Langmuir 2006, 22, 3728–3734. [Google Scholar] [CrossRef] [PubMed]
- Krueger, A.; Liang, Y.; Jarre, G.; Stegk, J. Surface functionalisation of detonation diamond suitable for biological applications. J. Mater. Chem. 2006, 16, 2322–2328. [Google Scholar] [CrossRef]
- Steenackers, M.; Lud, S.Q.; Niedermeier, M.; Bruno, P.; Gruen, D.M.; Feulner, P.; Stutzmann, M.; Garrido, J.A.; Jordan, R. Structured polymer grafts on diamond. J. Am. Chem. Soc. 2007, 129, 15655–15661. [Google Scholar] [CrossRef] [PubMed]
- Krueger, A.; Stegk, J.; Liang, Y.; Lu, L.; Jarre, G. Biotinylated nanodiamond: Simple and efficient functionalization of detonation diamond. Langmuir 2008, 24, 4200–4204. [Google Scholar] [CrossRef] [PubMed]
- Takáts, Z.; Wiseman, J.M.; Gologan, B.; Cooks, R.G. Mass spectrometry sampling under ambient conditions with desorption electrospray ionization. Science 2004, 306, 471–473. [Google Scholar] [CrossRef] [PubMed]
- McDonnell, L.A.; Heeren, R.M. Imaging mass spectrometry. Mass Spectrum. Rev. 2007, 26, 606–643. [Google Scholar] [CrossRef]
© 2010 by the authors. Licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Kong, X.; Cheng, P. Application of Nanodiamonds in Biomolecular Mass Spectrometry. Materials 2010, 3, 1845-1862. https://doi.org/10.3390/ma3031845
Kong X, Cheng P. Application of Nanodiamonds in Biomolecular Mass Spectrometry. Materials. 2010; 3(3):1845-1862. https://doi.org/10.3390/ma3031845
Chicago/Turabian StyleKong, Xianglei, and Ping Cheng. 2010. "Application of Nanodiamonds in Biomolecular Mass Spectrometry" Materials 3, no. 3: 1845-1862. https://doi.org/10.3390/ma3031845
APA StyleKong, X., & Cheng, P. (2010). Application of Nanodiamonds in Biomolecular Mass Spectrometry. Materials, 3(3), 1845-1862. https://doi.org/10.3390/ma3031845