Characteristics and Expression Analysis of FmTCP15 under Abiotic Stresses and Hormones and Interact with DELLA Protein in Fraxinus mandshurica Rupr.
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material and Growth Conditions
2.2. Cloning and Identification of FmTCP15 Gene
2.3. Sequence Features, Protein Modeling, and Phylogenetic Analysis of FmTCP15
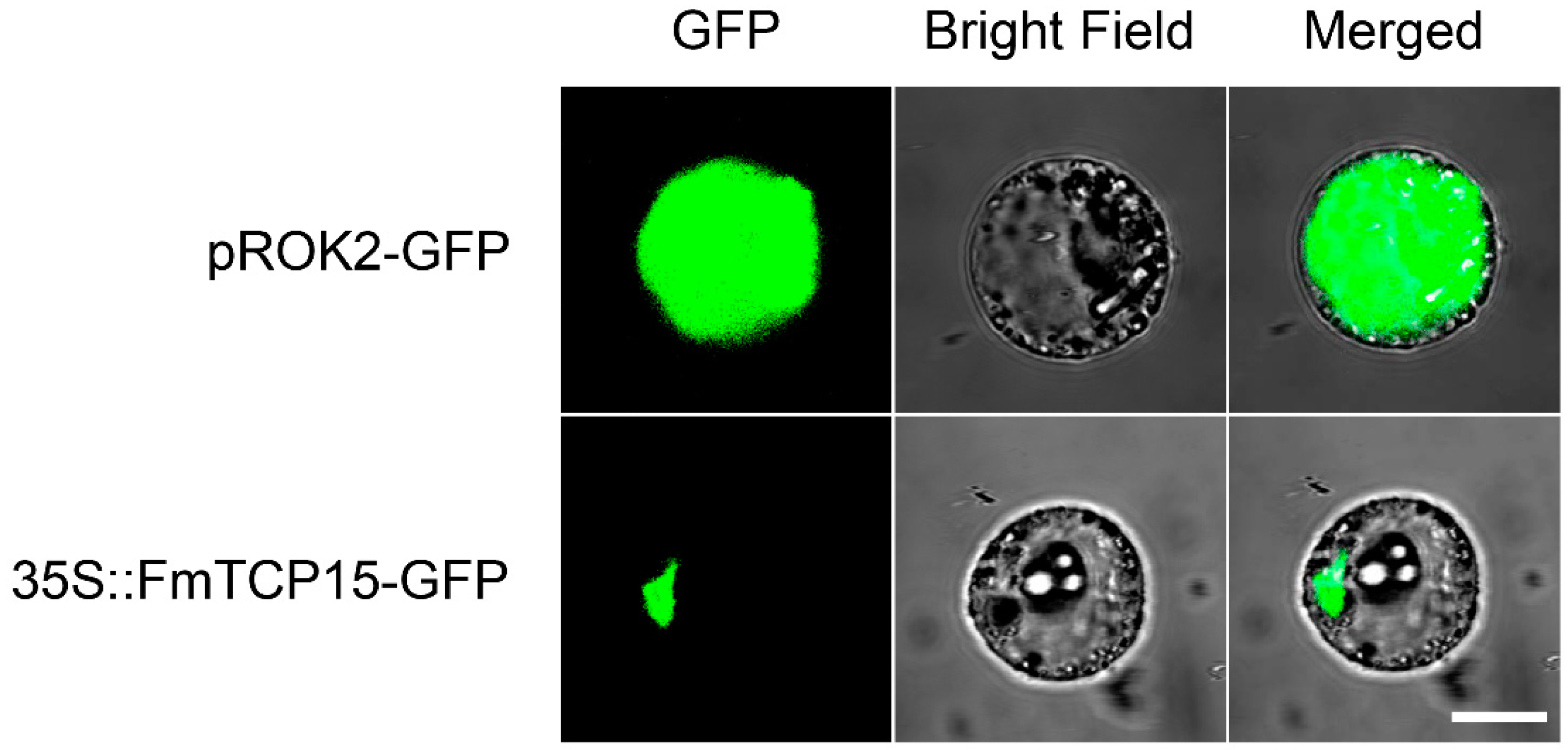
2.4. Subcellular Localization of FmTCP15 Proteins
2.5. Abiotic Stresses and Hormone Signal Treatments
2.6. Transient Overexpression of FmTCP15 Gene
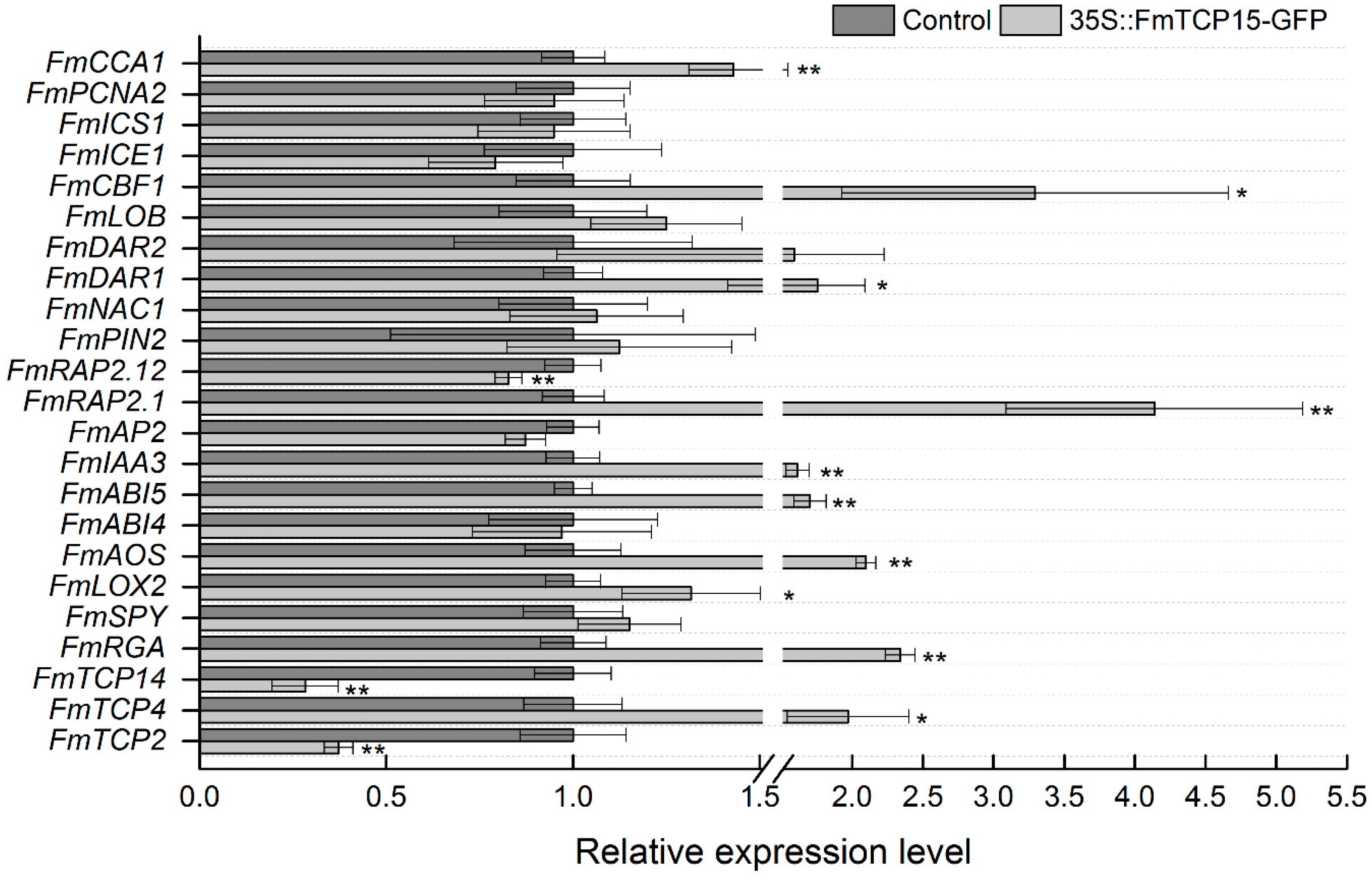
2.7. Analysis of Gene Expression of FmTCP15 and Downstream Genes
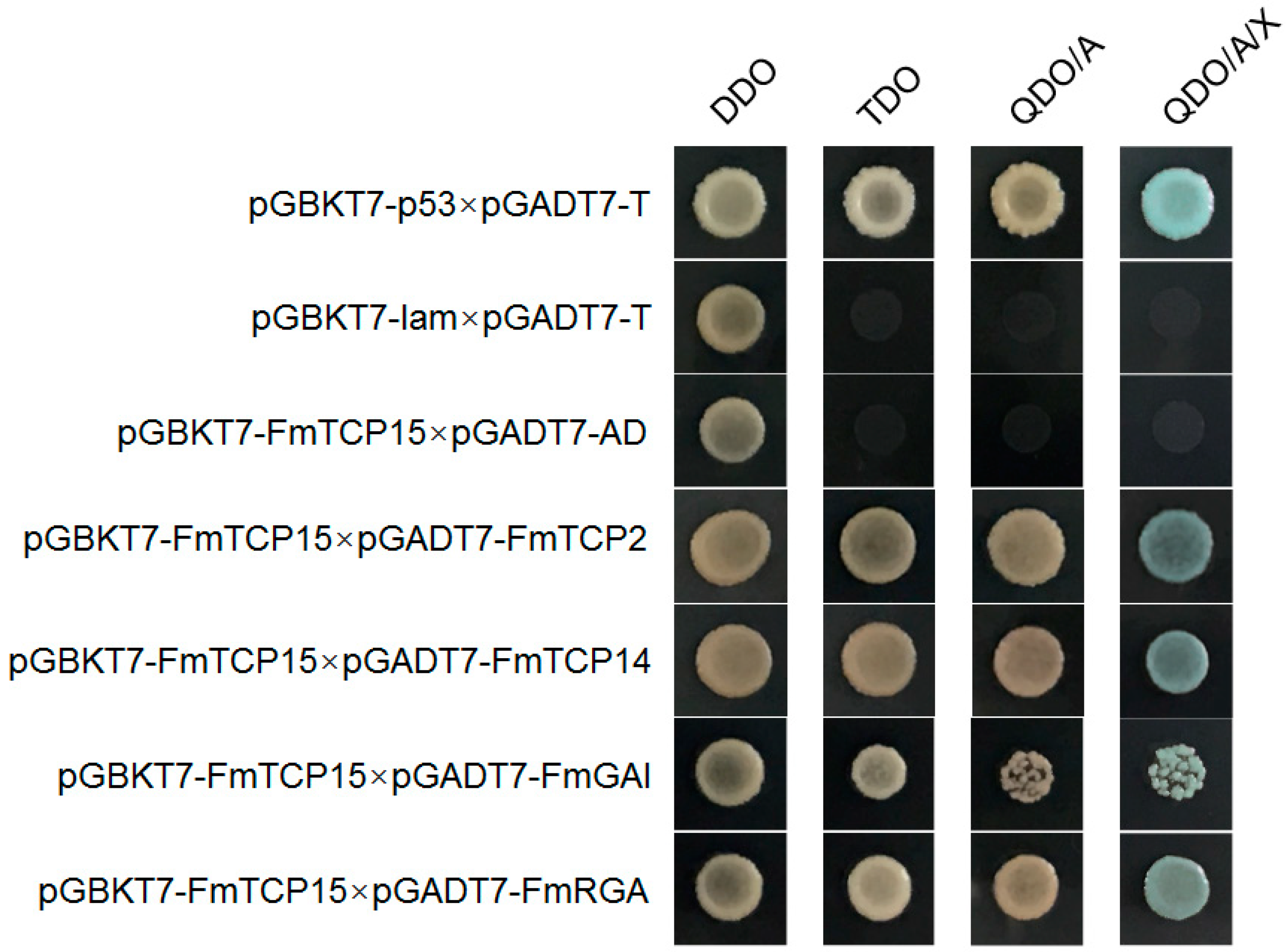
2.8. Yeast Two-Hybrid Protein–Protein Interaction Assays
3. Results
3.1. Nucleotide Sequence Cloning and Protein Modeling of FmTCP15 Gene
3.2. Homology Analysis and Phylogenetic Relationship of FmTCP15
3.3. Subcellular Localization of FmTCP15 Proteins
3.4. Expression Pattern of FmTCP15 in Different Tissues and Treatments
3.5. Functional Assay of Transient Overexpression of FmTCP15 in F. mandshurica
3.6. Protein–Protein Interactions between FmTCP15 and DELLA Proteins
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Luo, D.; Carpenter, R.; Vincent, C.; Copsey, L.; Coen, E. Origin of floral asymmetry in Antirrhinum. Nature 1996, 383, 794–799. [Google Scholar] [CrossRef] [PubMed]
- Doebley, J.; Stec, A.; Hubbard, L. The evolution of apical dominance in maize. Nature 1997, 386, 485–488. [Google Scholar] [CrossRef] [PubMed]
- Cubas, P.; Lauter, N.; Doebley, J.; Coen, E. The TCP domain: A motif found in proteins regulating plant growth and development. Plant J. 1999, 18, 215–222. [Google Scholar] [CrossRef] [PubMed]
- Kosugi, S.; Ohashi, Y. PCF1 and PCF2 specifically bind to cis elements in the rice proliferating cell nuclear antigen gene. Plant Cell. 1997, 9, 1607–1619. [Google Scholar] [CrossRef] [PubMed]
- Dhaka, N.; Bhardwaj, V.; Sharma, M.K.; Sharma, R. Evolving Tale of TCPs: New Paradigms and Old Lacunae. Front Plant Sci. 2017, 8, 479. [Google Scholar] [CrossRef] [PubMed]
- Kosugi, S.; Ohashi, Y. DNA binding and dimerization specificity and potential targets for the TCP protein family. Plant J. 2002, 30, 337–348. [Google Scholar] [CrossRef]
- Martín-Trillo, M.; Cubas, P. TCP genes: A family snapshot ten years later. Trends Plant Sci. 2010, 15, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Navaud, O.; Dabos, P.; Carnus, E.; Tremousaygue, D.; Hervé, C. TCP transcription factors predate the emergence of land plants. J. Mol. Evol. 2007, 65, 23–33. [Google Scholar] [CrossRef]
- Girardi, C.L.; Rombaldi, C.V.; Dal Cero, J.; Nobile, P.M.; Laurens, F.; Bouzayen, M.; Quecini, V. Genome-wide analysis of the AP2/ERF superfamily in apple and transcriptional evidence of ERF involvement in scab pathogenesis. Sci. Hortic. 2013, 151, 112–121. [Google Scholar] [CrossRef]
- Howarth, D.G.; Donoghue, M.J. Phylogenetic analysis of the “ECE”(CYC/TB1) clade reveals duplications predating the core eudicots. Proc. Natl. Acad. Sci. USA 2006, 103, 9101–9106. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Potuschak, T.; Colón-Carmona, A.; Gutiérrez, R.A.; Doerner, P. Arabidopsis TCP20 links regulation of growth and cell division control pathways. Proc. Natl. Acad. Sci. USA 2005, 102, 12978–12983. [Google Scholar] [CrossRef] [PubMed]
- Almeida, D.M.; Gregorio, G.B.; Oliveira, M.M.; Saibo, N.J. Five novel transcription factors as potential regulators of OsNHX1 gene expression in a salt tolerant rice genotype. Plant Mol. Biol. 2017, 93, 61–77. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, P.; Tyagi, A.K.; Tyagi, A.K. OsTCP19 influences developmental and abiotic stress signaling by modulating ABI4-mediated pathways. Sci. Rep. 2015, 5, 9998. [Google Scholar] [CrossRef]
- Palatnik, J.F.; Allen, E.; Wu, X.; Schommer, C.; Schwab, R.; Carrington, J.C.; Weigel, D. Control of leaf morphogenesis by microRNAs. Nature 2003, 425, 257–263. [Google Scholar] [CrossRef]
- Koyama, T.; Furutani, M.; Tasaka, M.; Ohme-Takagi, M. TCP transcription factors control the morphology of shoot lateral organs via negative regulation of the expression of boundary-specific genes in Arabidopsis. Plant Cell. 2007, 19, 473–484. [Google Scholar] [CrossRef]
- Schommer, C.; Palatnik, J.F.; Aggarwal, P.; Chételat, A.; Cubas, P.; Farmer, E.E.; Nath, U.; Weigel, D. Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol. 2008, 6, e230. [Google Scholar] [CrossRef]
- Yang, X.; Zhao, X.; Li, C.; Liu, J.; Qiu, Z.; Dong, Y.; Wang, Y. Distinct regulatory changes underlying differential expression of TEOSINTE BRANCHED1-CYCLOIDEA-PROLIFERATING CELL FACTOR genes associated with petal variations in zygomorphic flowers of Petrocosmea spp. of the family Gesneriaceae. Plant Physiol. 2015, 169, 2138–2151. [Google Scholar] [PubMed]
- Gonzalezgrandio, E.; Pozacarrion, C.; Oscar, C.; Sorzano, S.; Cubas, P. BRANCHED1 Promotes Axillary Bud Dormancy in Response to Shade in Arabidopsis. Plant Cell. 2013, 25, 834–850. [Google Scholar] [CrossRef]
- Reddy, S.K.; Holalu, S.V.; Casal, J.J.; Finlayson, S.A. Abscisic acid regulates axillary bud outgrowth responses to the ratio of red to far-red light. Plant Physiol. 2013, 163, 1047–1058. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Xu, Y.; Liu, H.; Mao, Z.; Zhang, C.; Ma, Y.; Zhang, Q.; Meng, Z.; Chong, K. The interaction between OsMADS57 and OsTB1 modulates rice tillering via DWARF14. Nat. Commun. 2013, 4, 1566. [Google Scholar] [CrossRef]
- Costa, M.M.R.; Fox, S.; Hanna, A.I.; Baxter, C.; Coen, E. Evolution of regulatory interactions controlling floral asymmetry. Development 2005, 132, 5093–5101. [Google Scholar] [CrossRef] [PubMed]
- Danisman, S.; Van der Wal, F.; Dhondt, S.; Waites, R.; de Folter, S.; Bimbo, A.; van Dijk, A.D.; Muino, J.M.; Cutri, L.; Dornelas, M.C. Arabidopsis class I and class II TCP transcription factors regulate jasmonic acid metabolism and leaf development antagonistically. Plant Physiol. 2012, 159, 1511–1523. [Google Scholar] [CrossRef]
- Parapunova, V.; Busscher, M.; Busscher-Lange, J.; Lammers, M.; Karlova, R.; Bovy, A.G.; Angenent, G.C.; de Maagd, R.A. Identification, cloning and characterization of the tomato TCP transcription factor family. BMC Plant Biol. 2014, 14, 157. [Google Scholar] [CrossRef] [PubMed]
- Davière, J.M.; Wild, M.; Regnault, T.; Baumberger, N.; Eisler, H.; Genschik, P.; Achard, P. Class I TCP-DELLA interactions in inflorescence shoot apex determine plant height. Curr. Biol. 2014, 24, 1923–1928. [Google Scholar] [CrossRef] [PubMed]
- Torres, M.L.D.L.Y.; Palomares, O.; Quiralte, J.; Pauli, G.; Rodriguez, R.; Villalba, M. An Enzymatically Active β-1,3-Glucanase from Ash Pollen with Allergenic Properties: A Particular Member in the Oleaceae Family. PLoS ONE 2015, 10, e0133066. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Yang, D.; Niu, C.; Zhang, W.; Wang, M.; Hao, G. Correlation between leaf size and hydraulic architecture in five compound-leaved tree species of a temperate forest in NE China. For. Ecol. Manage. 2017, 418, 63–72. [Google Scholar] [CrossRef]
- Gang, Q.; Yan, Q.; Zhu, J. Effects of thinning on early seed regeneration of two broadleaved tree species in larch plantations: Implication for converting pure larch plantations into larch-broadleaved mixed forests. Forestry 2015, 88, 573–585. [Google Scholar] [CrossRef]
- Huang, S.; Sun, X.; Zhang, Y.; Sun, H.; Wang, Z. Nutrient retranslocation from the fine roots of Fraxinus mandshurica and Larix olgensis in northeastern China. J. For. Res. 2016, 27, 1305–1312. [Google Scholar] [CrossRef]
- Li, T.; Wu, J.; Chen, H.; Ji, L.; Yu, D.; Zhou, L.; Zhou, W.; Tong, Y.; Li, Y.; Dai, L. Intraspecific functional trait variability across different spatial scales: A case study of two dominant trees in Korean pine broadleaved forest. Plant Ecol. 2018, 219, 875–886. [Google Scholar] [CrossRef]
- Drenkhan, R.; Solheim, H.; Bogacheva, A.M.; Riit, T.; Adamson, K.; Drenkhan, T.; Maaten, T.; Hietala, A.M. Hymenoscyphus fraxineus is a leaf pathogen of local Fraxinus species in the Russian Far East. Plant Pathol. 2017, 66, 490–500. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.e.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools on the ExPASy server. In The Proteomics Protocols Handbook, 1st ed.; John, M.W., Ed.; Humana Press: Clifton, NJ, USA, 2005; pp. 571–607. [Google Scholar]
- Hofmann, K. TMbase-A database of membrane spanning proteins segments. Biol. Chem. Hoppe-Seyler. 1993, 374, 166. [Google Scholar]
- Wang, Y.; Virtanen, J.; Xue, Z.; Tesmer, J.J.; Zhang, Y. Using iterative fragment assembly and progressive sequence truncation to facilitate phasing and crystal structure determination of distantly related proteins. Acta Crystallogr. D 2016, 72, 616–628. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Virtanen, J.; Xue, Z.; Zhang, Y. I-TASSER-MR: Automated molecular replacement for distant-homology proteins using iterative fragment assembly and progressive sequence truncation. Nucleic Acids Res. 2017, 45, 429–434. [Google Scholar] [CrossRef]
- Xu, D.; Zhang, Y. Improving the physical realism and structural accuracy of protein models by a two-step atomic-level energy minimization. Biophys. J. 2011, 101, 2525–2534. [Google Scholar] [CrossRef]
- Lin, Y.; Li, W.; Chen, H.; Li, Q.; Sun, Y.; Shi, R.; Lin, C.; Wang, J.P.; Chen, H.; Chuang, L. A simple improved-throughput xylem protoplast system for studying wood formation. Nat. Protoc. 2014, 9, 2194–2205. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, Y.; Wang, C. Gene overexpression and gene silencing in Birch using an Agrobacterium-mediated transient expression system. Mol. Biol. Rep. 2012, 39, 5537–5541. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Kieffer, M.; Master, V.; Waites, R.; Davies, B. TCP14 and TCP15 affect internode length and leaf shape in Arabidopsis. Plant J. 2011, 68, 147–158. [Google Scholar] [CrossRef] [PubMed]
- Resentini, F.; Felipo-Benavent, A.; Colombo, L.; Blázquez, M.A.; Alabadí, D.; Masiero, S. TCP14 and TCP15 mediate the promotion of seed germination by gibberellins in Arabidopsis thaliana. Mol. Plant. 2015, 8, 482–485. [Google Scholar] [CrossRef]
- Lei, N.; Yu, X.; Li, S.; Zeng, C.; Zou, L.; Liao, W.; Peng, M. Phylogeny and expression pattern analysis of TCP transcription factors in cassava seedlings exposed to cold and/or drought stress. Sci. Rep. 2017, 7, 10016. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Zhai, Z.; Li, Y.; Geng, S.; Song, G.; Guan, J.; Jia, M.; Wang, F.; Sun, G.; Feng, N. Genome-Wide Identification and Expression Profiling of the TCP Family Genes in Spike and Grain Development of Wheat (Triticum aestivum L.). Front Plant Sci. 2018, 9, 1282. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Zhao, P.; Cheng, H.; Han, L.; Wu, X.; Gao, P.; Wang, H.; Yang, C.; Zhong, N.; Zuo, J. The cotton transcription factor TCP14 functions in auxin-mediated epidermal cell differentiation and elongation. Plant Physiol. 2013, 162, 1669–1680. [Google Scholar] [CrossRef]
- Tian, Q.; Uhlir, N.J.; Reed, J.W. Arabidopsis SHY2/IAA3 inhibits auxin-regulated gene expression. Plant Cell. 2002, 14, 301–319. [Google Scholar] [CrossRef] [PubMed]
- Mao, D.; Chen, C. Colinearity and similar expression pattern of rice DREB1s reveal their functional conservation in the cold-responsive pathway. PLoS ONE 2012, 7, e47275. [Google Scholar] [CrossRef]
- Wu, J.; Tsai, H.; Joanito, I.; Wu, Y.; Chang, C.; Li, Y.; Wang, Y.; Hong, J.C.; Chu, J.; Hsu, C.; et al. LWD–TCP complex activates the morning gene CCA1 in Arabidopsis. Nat. Commun. 2016, 7, 13181. [Google Scholar] [CrossRef]
- Colebrook, E.H.; Thomas, S.G.; Phillips, A.; Hedden, P. The role of gibberellin signalling in plant responses to abiotic stress. J. Exp. Biol. 2014, 217, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; De Storme, N.; Geelen, D. Cold-Induced Male Meiotic Restitution in Arabidopsis thaliana Is Not Mediated by GA-DELLA Signaling. Front Plant Sci. 2018, 9, 91. [Google Scholar] [CrossRef] [PubMed]
- Achard, P.; Gong, F.; Cheminant, S.; Alioua, M.; Hedden, P.; Genschik, P. The Cold-Inducible CBF1 Factor–Dependent Signaling Pathway Modulates the Accumulation of the Growth-Repressing DELLA Proteins via Its Effect on Gibberellin Metabolism. Plant Cell. 2008, 20, 2117–2129. [Google Scholar] [CrossRef]
- King, K.E.; Moritz, T.; Harberd, N.P. Gibberellins Are Not Required for Normal Stem Growth in Arabidopsis thaliana in the Absence of GAI and RGA. Genetics 2001, 159, 767–776. [Google Scholar] [PubMed]
- Yang, D.L.; Yao, J.; Mei, C.S.; Tong, X.H.; Zeng, L.J.; Li, Q.; Xiao, L.T.; Sun, T.; Li, J.; Deng, X.W. Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc. Natl. Acad. Sci. USA 2012, 109, 7152–7153. [Google Scholar] [CrossRef]
- Verma, V.; Ravindran, P.; Kumar, P.P. Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 2016, 16, 86. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Chen, H.; Wei, D.; Ma, H.; Lin, J. Arabidopsis CBF3 and DELLAs positively regulate each other in response to low temperature. Sci. Rep. 2017, 7, 39819. [Google Scholar] [CrossRef] [PubMed]



© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liang, N.; Zhan, Y.; Yu, L.; Wang, Z.; Zeng, F. Characteristics and Expression Analysis of FmTCP15 under Abiotic Stresses and Hormones and Interact with DELLA Protein in Fraxinus mandshurica Rupr. Forests 2019, 10, 343. https://doi.org/10.3390/f10040343
Liang N, Zhan Y, Yu L, Wang Z, Zeng F. Characteristics and Expression Analysis of FmTCP15 under Abiotic Stresses and Hormones and Interact with DELLA Protein in Fraxinus mandshurica Rupr. Forests. 2019; 10(4):343. https://doi.org/10.3390/f10040343
Chicago/Turabian StyleLiang, Nansong, Yaguang Zhan, Lei Yu, Ziqing Wang, and Fansuo Zeng. 2019. "Characteristics and Expression Analysis of FmTCP15 under Abiotic Stresses and Hormones and Interact with DELLA Protein in Fraxinus mandshurica Rupr." Forests 10, no. 4: 343. https://doi.org/10.3390/f10040343
APA StyleLiang, N., Zhan, Y., Yu, L., Wang, Z., & Zeng, F. (2019). Characteristics and Expression Analysis of FmTCP15 under Abiotic Stresses and Hormones and Interact with DELLA Protein in Fraxinus mandshurica Rupr. Forests, 10(4), 343. https://doi.org/10.3390/f10040343




