Study on Gene Differential Expression in Tetraploid Populus Leaves
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. Measurement of Leaf Areas
2.3. Determination of the Chlorophyll Content
2.4. Determination of the Auxin Content
2.5. RNA Extraction, RNA-seq Library Preparation, and Sequencing
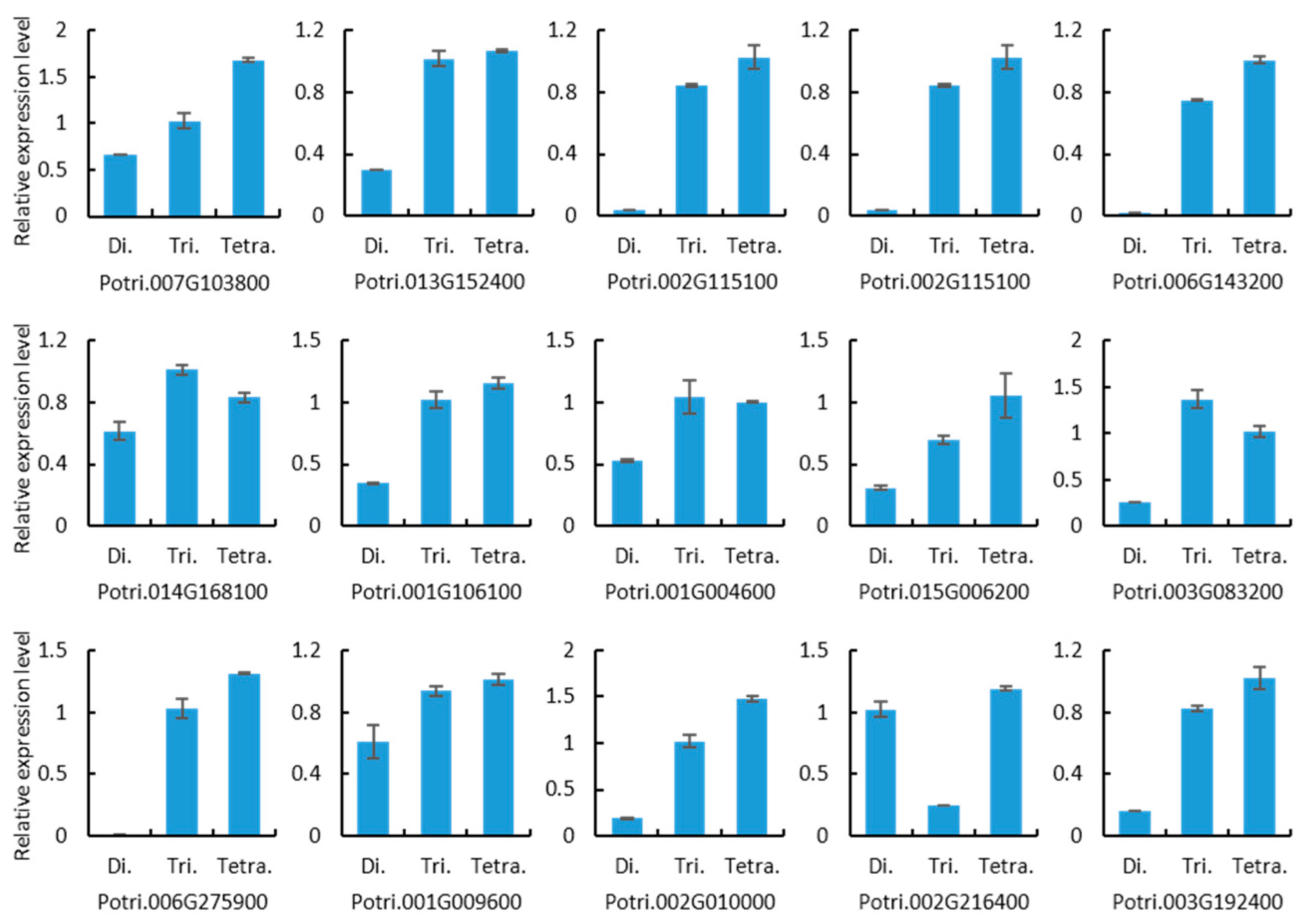
2.6. Quantitative Real-Time PCR Analysis of Gene Expression
2.7. Statistical Analysis
3. Results
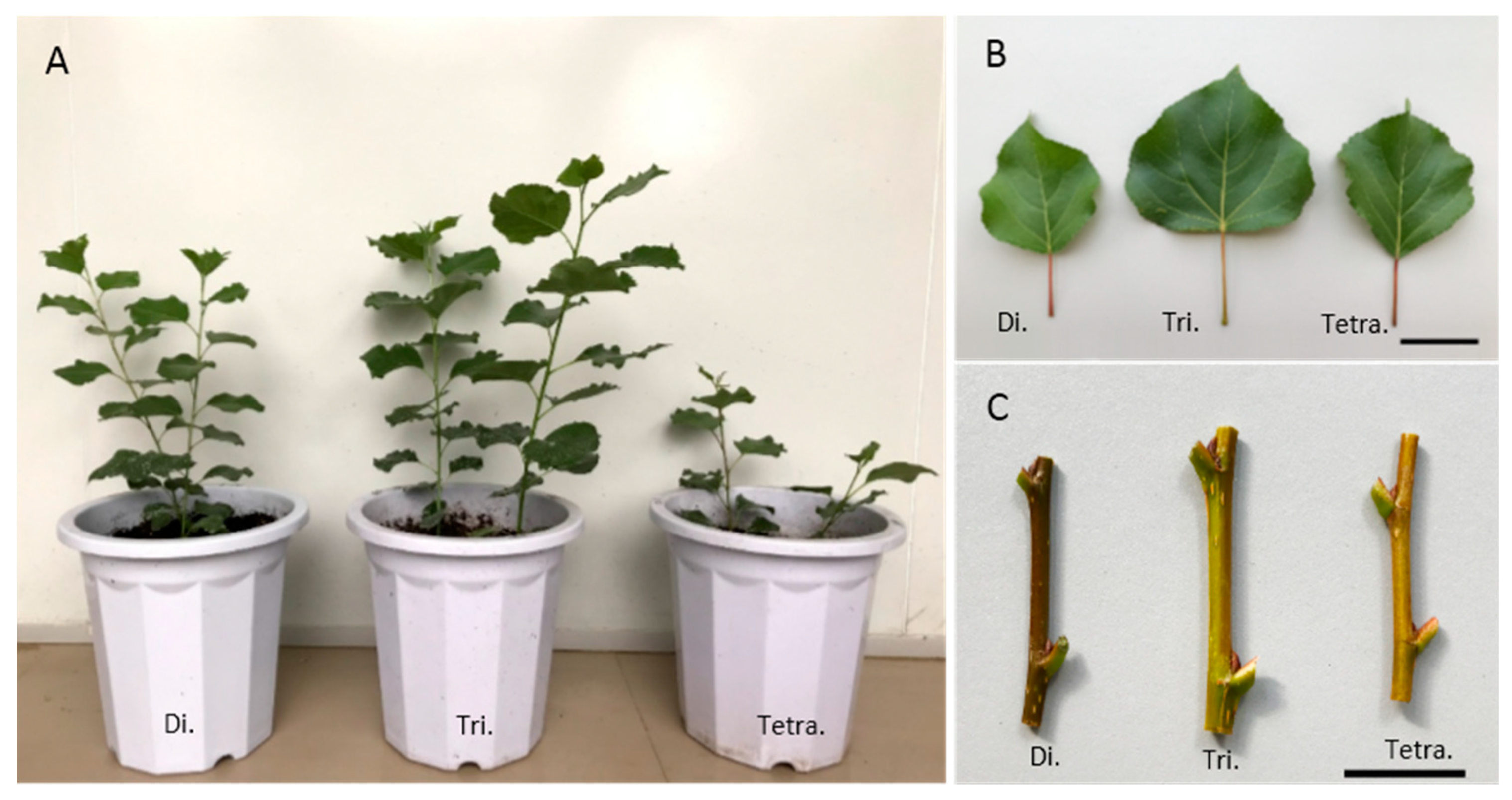
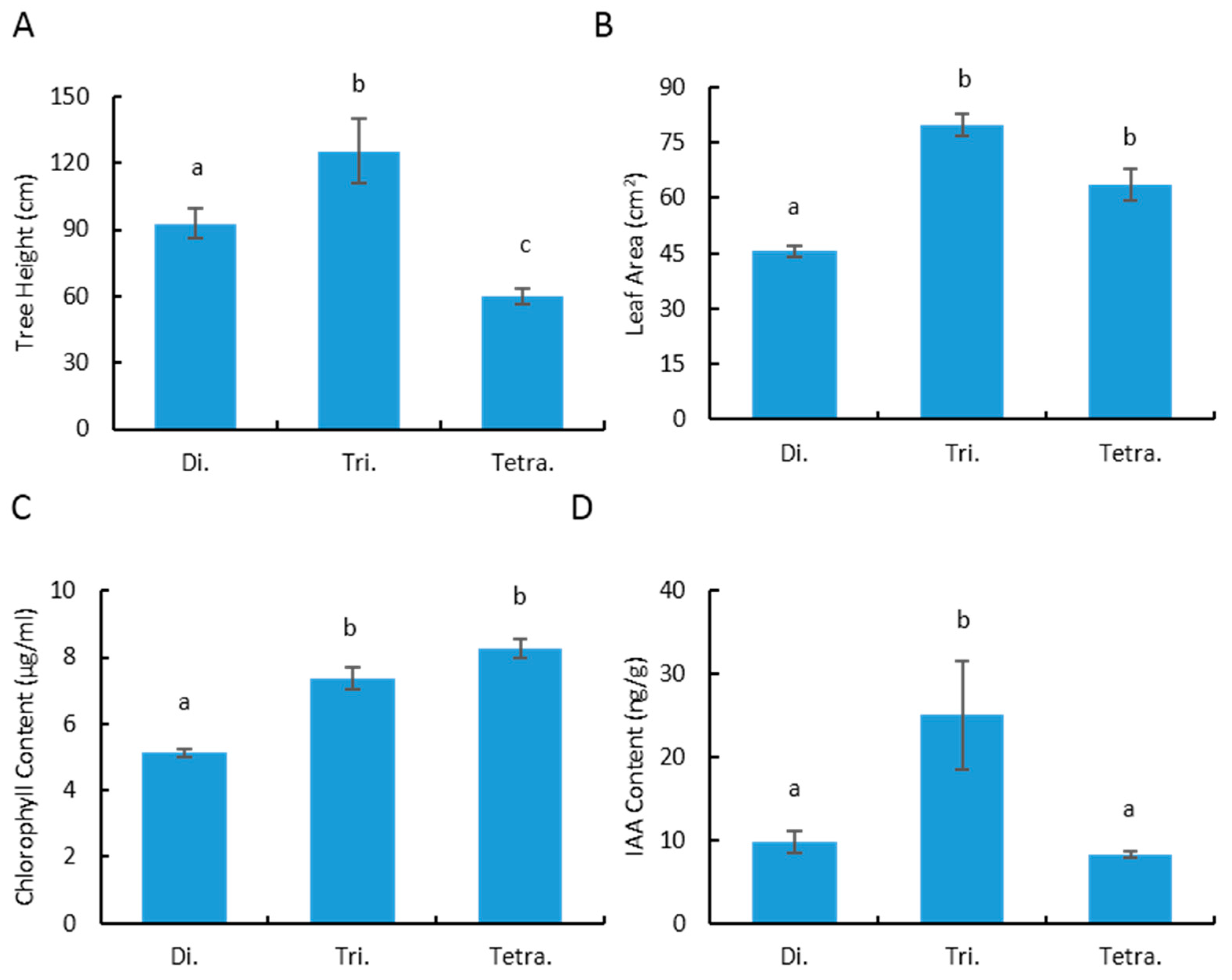
3.1. Variation of the Polyploidy Plant Growth, Leaf Morphology and Physiological Characteristic in Poplar
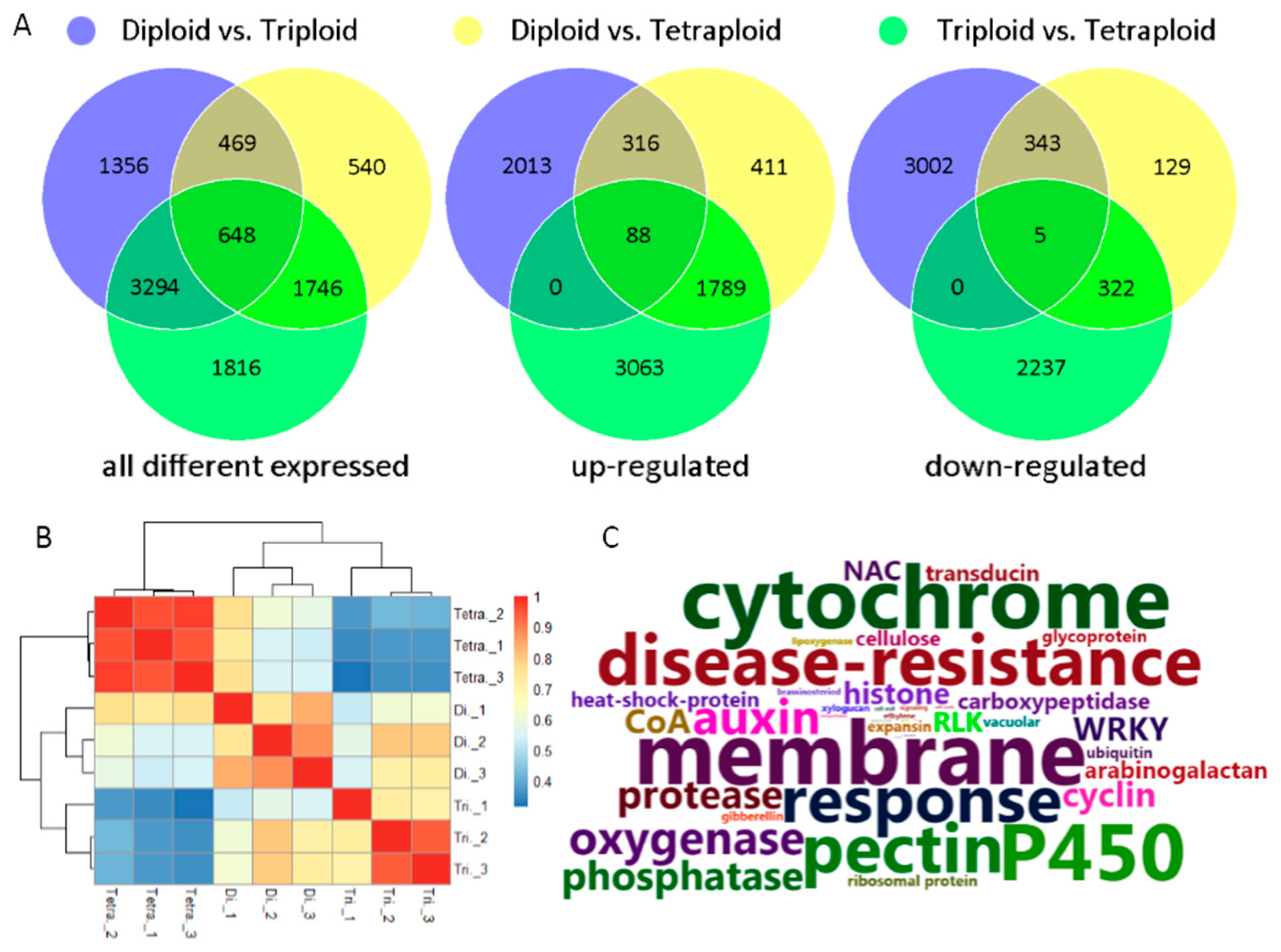
3.2. Gene Expressions and Differentially Expressed Genes (DEGs) among Tetraploid, Triploid, and Diploid
3.3. Analysis of the Differential Expression and Functional Enrichment of Leaf Genes in Poplar Tetraploid and Diploid
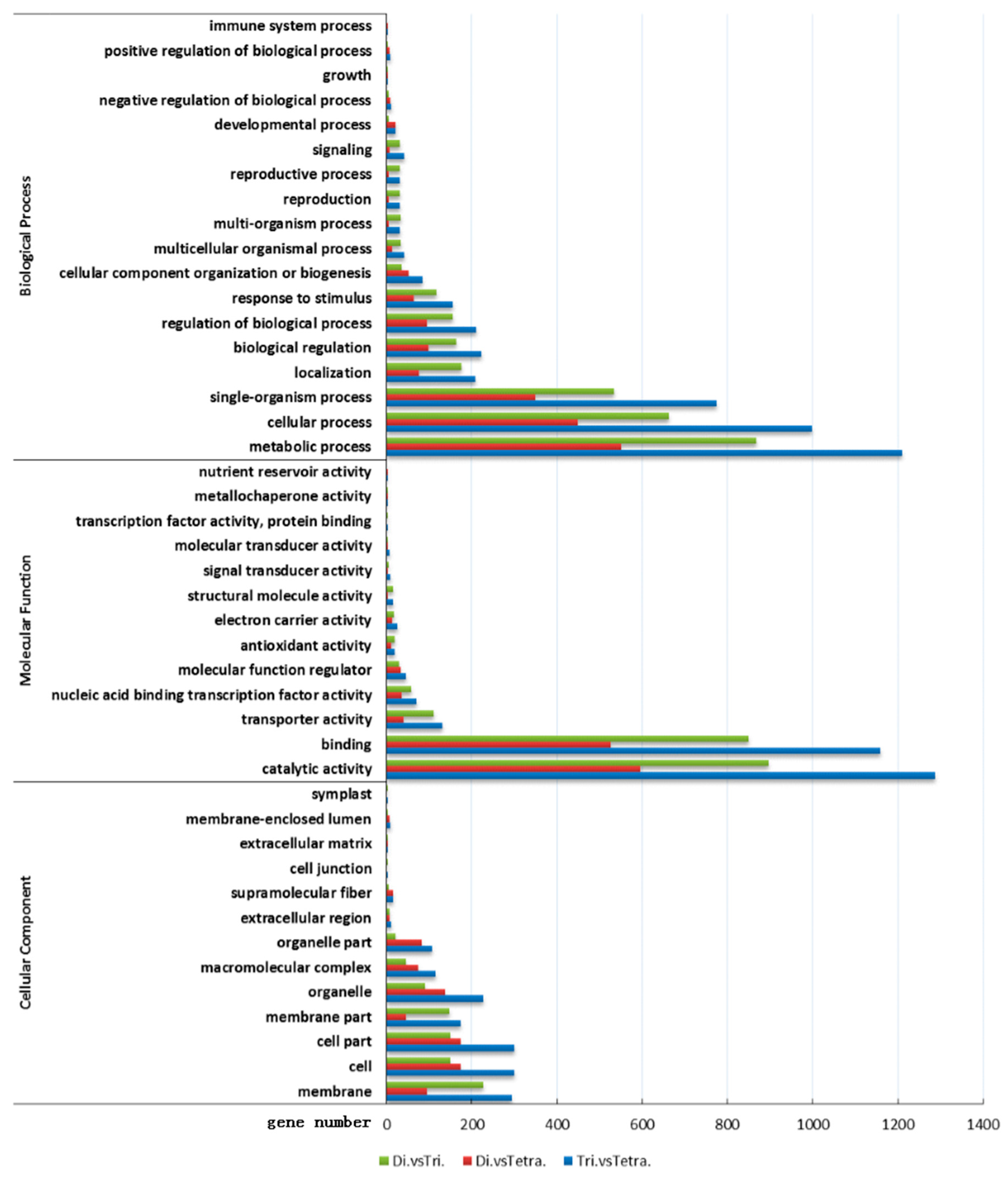
3.3.1. GO Enrichment Analysis of Differential Genes in Poplar Tetraploid and Diploid Leaves
3.3.2. KEGG Enrichment Analysis of Differential Genes in Poplar Tetraploid and Diploid Leaves
3.4. Analysis of the Differential Expression and Functional Enrichment of Leaf Genes in Poplar Tetraploid and Triploid
3.4.1. GO Enrichment Analysis of Differential Genes in Poplar Tetraploid and Triploid Leaves
3.4.2. KEGG Enrichment Analysis of Differential Genes in Poplar Tetraploid and Triploid Leaves
4. Discussion
4.1. Differential Expression of Genes Related to Leaf Growth and Chlorophyll Synthesis of Tetraploid Poplar
4.2. Differential Expression of Auxin and Senescence-Related Genes in Tetraploid Poplar Leaves
4.3. Differential Expression of Genes Related to Chlorophyll Synthesis in Tetraploid Poplar Leaves
4.4. Differential Expression of Genes Related to Photosynthesis and Carbon Fixation in Tetraploid Poplar Leaves
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Leitch, A.R.; Leitch, I.J. Perspective—Genomic plasticity and the diversity of polyploid plants. Science 2008, 320, 481–483. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.J. Molecular mechanisms of polyploidy and hybrid vigor. Trends Plant Sci. 2010, 15, 57–71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ng, D.W.-K.; Lu, J.; Chen, Z.J. Big roles for small RNAs in polyploidy, hybrid vigor, and hybrid incompatibility. Curr. Opin. Plant Biol. 2012, 15, 154–161. [Google Scholar] [CrossRef] [PubMed]
- Nilsson-Ehle, H. Note regarding the gigas form of Populus tremula found in nature. Hereditas 1936, 21, 372–382. [Google Scholar]
- Zhu, Z.; Lin, H.; Kang, X. Studies on allotriploid breeding of Populus tomentosa B301 clones. Sci. Silvae Sin. 1995, 31, 499–506. [Google Scholar]
- Kondorosi, E.; Roudier, F.; Gendreau, E. Plant cell-size control: Growing by ploidy? Curr. Opin. Plant Biol. 2000, 3, 488–492. [Google Scholar] [CrossRef]
- Ni, Z.; Kim, E.-D.; Ha, M.; Lackey, E.; Liu, J.; Zhang, Y.; Sun, Q.; Chen, Z.J. Altered circadian rhythms regulate growth vigour in hybrids and allopolyploids. Nat. Cell Biol. 2009, 457, 327–331. [Google Scholar] [CrossRef] [Green Version]
- Miller, M.E.; Zhang, C.; Chen, Z.J. Ploidy and Hybridity Effects on Growth Vigor and Gene Expression in Arabidopsis thaliana Hybrids and Their Parents. G3 Genes Genomes Genet. 2012, 2, 505–513. [Google Scholar] [CrossRef] [Green Version]
- Xiang, D.; Quilichini, T.D.; Liu, Z.; Gao, P.; Pan, Y.; Li, Q.; Nilsen, K.T.; Venglat, P.; Esteban, E.; Pasha, A.; et al. The Transcriptional Landscape of Polyploid Wheats and Their Diploid Ancestors during Embryogenesis and Grain Development. Plant Cell 2019, 31, 2888–2911. [Google Scholar] [CrossRef] [Green Version]
- Shewry, P.R. Wheat. J. Exp. Bot. 2009, 60, 1537–1553. [Google Scholar] [CrossRef]
- Jiang, C.-X.; Wright, R.J.; El-Zik, K.M.; Paterson, A.H. Polyploid formation created unique avenues for response to selection in Gossypium (cotton). Proc. Natl. Acad. Sci. USA 1998, 95, 4419–4424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allario, T.; Brumos, J.; Colmenero-Flores, J.M.; Tadeo, F.; Froelicher, Y.; Talon, M.; Navarro, L.; Ollitrault, P.; Morillon, R. Large changes in anatomy and physiology between diploid Rangpur lime (Citrus limonia) and its autotetraploid are not associated with large changes in leaf gene expression. J. Exp. Bot. 2011, 62, 2507–2519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mu, H.-Z.; Liu, Z.-J.; Lin, L.; Li, H.-Y.; Jiang, J.; Liu, G. Transcriptomic Analysis of Phenotypic Changes in Birch (Betula platyphylla) Autotetraploids. Int. J. Mol. Sci. 2012, 13, 13012–13029. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Estrada-Johnson, E.; Csukasi, F.; Pizarro, C.M.; Vallarino, J.G.; Kiryakova, Y.; Vioque, A.; Brumos, J.; Medina-Escobar, N.; Botella, M.A.; Alonso, J.M.; et al. Transcriptomic Analysis in Strawberry Fruits Reveals Active Auxin Biosynthesis and Signaling in the Ripe Receptacle. Front. Plant Sci. 2017, 8, 889. [Google Scholar] [CrossRef] [Green Version]
- Saminathan, T.; Nimmakayala, P.; Manohar, S.; Malkaram, S.; Almeida, A.; Cantrell, R.; Tomason, Y.; Abburi, L.; Rahman, M.A.; Vajja, V.G.; et al. Differential gene expression and alternative splicing between diploid and tetraploid watermelon. J. Exp. Bot. 2015, 66, 1369–1385. [Google Scholar] [CrossRef]
- Stupar, R.M.; Bhaskar, P.B.; Yandell, B.S.; Rensink, W.A.; Hart, A.L.; Ouyang, S.; Veilleux, R.E.; Busse, J.S.; Erhardt, R.J.; Buell, C.R.; et al. Phenotypic and Transcriptomic Changes Associated With Potato Autopolyploidization. Genetics 2007, 176, 2055–2067. [Google Scholar] [CrossRef] [Green Version]
- Albertin, W.; Brabant, P.; Catrice, O.; Eber, F.; Jenczewski, E.; Chèvre, A.M.; Thiellement, H. Autopolyploidy in cabbage (Brassica oleracea L.) does not alter significantly the proteomes of green tissues. Proteomics 2005, 5, 2131–2139. [Google Scholar] [CrossRef]
- Liao, T.; Cheng, S.; Zhu, X.; Min, Y.; Kang, X. Effects of triploid status on growth, photosynthesis, and leaf area in Populus. Trees 2016, 30, 1137–1147. [Google Scholar] [CrossRef]
- Cheng, S. Gene expression changes in the full-sib allotriploid population of Populus spp. (Section Tacamahaca). Euphytica 2015, 203, 683–700. [Google Scholar] [CrossRef]
- Suo, Y.; Min, Y.; Dong, C.; Wang, Y.; Cheng, S.; Kang, X. MicroRNA expression changes following synthesis of three full-sib Populus triploid populations with different heterozygosities. Plant Mol. Biol. 2017, 95, 215–225. [Google Scholar] [CrossRef]
- Du, K.; Liao, T.; Ren, Y.; Geng, X.; Kang, X. Molecular mechanism of vegetative growth advantage in allotriploid Populus. Int. J. Mol. Sci. 2020, 21, 441. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, K.; Han, Q.; Zhang, Y.; Kang, X. Differential Expression of Genes Related to the Formation of Giant Leaves in Triploid Poplar. Forests 2019, 10, 920. [Google Scholar] [CrossRef] [Green Version]
- Congping, X. Tetraploid induction and molecular basic of vegetative slowly growth in tetrapoid Populus spp. (Section Tcamahaca). Silva Fenn. 2018, 47, 2. [Google Scholar]
- Dong, C.-B.; Suo, Y.-J.; Kang, X.-Y. Assessment of the genetic composition of triploid hybrid Populus using SSR markers with low recombination frequencies. Can. J. For. Res. 2014, 44, 692–699. [Google Scholar] [CrossRef]
- Xu, C.; Huang, Z.; Liao, T.; Li, Y.; Kang, X. In vitro tetraploid plants regeneration from leaf explants of multiple genotypes in Populus. Plant Cell Tissue Organ Cult. (PCTOC) 2015, 125, 1–9. [Google Scholar] [CrossRef]
- Lichtenthalermpresidentoffespp, H.; Buschmann, C. Chlorophylls and Carotenoids: Measurement and Characterization by UV-VIS Spectroscopy. Curr. Protoc. Food Anal. Chem. 2001, 1, F4.3.1–F4.3.8. [Google Scholar] [CrossRef]
- Novák, O.; Hényková, E.; Sairanen, I.; Kowalczyk, M.; Pospíšil, T.; Ljung, K. Tissue-specific profiling of theArabidopsis thalianaauxin metabolome. Plant J. 2012, 72, 523–536. [Google Scholar] [CrossRef]
- Xiaojuan, S.; Baoguo, F.; Lichai, Y.; XiuNa, C.; Shanfa, L. Selection and Validation of Reference Genes for Quantitative RT-PCR Analysis of Gene Expression in Populus trichocarpa. Chin. Bull. Bot. 2014, 48, 507–518. [Google Scholar] [CrossRef]
- Zhang, L.; Zou, J.; Li, S.; Wang, B.; Raboanatahiry, N.; Li, M. Characterization and expression profiles of miRNAs in the triploid hybrids of Brassica napus and Brassica rapa. BMC Genom. 2019, 20, 649. [Google Scholar] [CrossRef] [Green Version]
- Meng, Y.; Li, J.; Liu, J.; Hu, H.; Li, W.; Liu, W.; Chen, S. Ploidy effect and genetic architecture exploration of stalk traits using DH and its corresponding haploid populations in maize. BMC Plant Biol. 2016, 16, 50. [Google Scholar] [CrossRef] [Green Version]
- Lichtenthaler, H.K. Chlorophylls and Carotenoids: Pigments of Photosynthetic Biomembranes. Methods in Enzymology; Academic Press: Orlando, FL, USA, 1987; Volume 148, pp. 183–350. [Google Scholar]
- Heyes, D.J.; Hunter, C.N. Making light work of enzyme catalysis: Protochlorophyllide oxidoreductase. Trends Biochem. Sci. 2005, 30, 642–649. [Google Scholar] [CrossRef] [PubMed]
- Paddock, T.; Mason, M.E.; Lima, D.F.; Armstrong, G.A. Arabidopsis protochlorophyllide oxidoreductase A (PORA) restores bulk chlorophyll synthesis and normal development to a porB porC double mutant. Plant Mol. Biol. 2009, 72, 445–457. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.; Apel, K. Arabidopsis light-dependent NADPH: Protochlorophyllide oxidoreductase A (PORA) is essential for normal plant growth and development: An addendum. Plant Mol. Biol. 2012, 80, 237–240. [Google Scholar] [CrossRef] [PubMed]
- Reyes, J.C.; Muro-Pastor, M.I.; Florencio, F.J. The GATA Family of Transcription Factors in Arabidopsis and Rice. Plant Physiol. 2004, 134, 1718–1732. [Google Scholar] [CrossRef] [Green Version]
- Bi, Y.-M.; Zhang, Y.; Signorelli, T.; Zhao, R.; Zhu, T.; Rothstein, S. Genetic analysis of Arabidopsis GATA transcription factor gene family reveals a nitrate-inducible member important for chlorophyll synthesis and glucose sensitivity. Plant J. 2005, 44, 680–692. [Google Scholar] [CrossRef]
- Hudson, D.; Guevara, D.R.; Hand, A.J.; Xu, Z.; Hao, L.; Chen, X.; Zhu, T.; Bi, Y.-M.; Rothstein, S.J. Rice Cytokinin GATA Transcription Factor1 Regulates Chloroplast Development and Plant Architecture. Plant Physiol. 2013, 162, 132–144. [Google Scholar] [CrossRef] [Green Version]
- An, Y.; Zhou, Y.; Han, X.; Shen, C.; Wang, S.; Liu, C.; Yin, W.; Xia, X. The GATA transcription factor GNC plays an important role in photosynthesis and growth in poplar. J. Exp. Bot. 2019, 71, 1969–1984. [Google Scholar] [CrossRef]
- Zhang, C.; Huang, Y.; Xiao, Z.; Yang, H.; Hao, Q.; Yuan, S.; Chen, H.; Chen, L.; Chen, S.; Zhou, X.; et al. A GATA Transcription Factor from Soybean (Glycine max) Regulates Chlorophyll Biosynthesis and Suppresses Growth in the Transgenic Arabidopsis thaliana. Plants 2020, 9, 1036. [Google Scholar] [CrossRef]
- Teale, W.D.; Paponov, I.; Palme, K. Auxin in action: Signalling, transport and the control of plant growth and development. Nat. Rev. Mol. Cell Biol. 2006, 7, 847–859. [Google Scholar] [CrossRef]
- Leyser, O. Auxin Signaling. Plant Physiol. 2018, 176, 465–479. [Google Scholar] [CrossRef] [Green Version]
- Mroue, S.; Simeunovic, A.; Robert, H.S. Auxin production as an integrator of environmental cues for developmental growth regulation. J. Exp. Bot. 2018, 69, 201–212. [Google Scholar] [CrossRef] [PubMed]
- Woodward, A.W. Auxin: Regulation, Action, and Interaction. Ann. Bot. 2005, 95, 707–735. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- E Staswick, P.; Serban, B.; Rowe, M.; Tiryaki, I.; Maldonado, M.T.; Maldonado, M.C.; Suza, W. Characterization of an Arabidopsis Enzyme Family That Conjugates Amino Acids to Indole-3-Acetic Acid. Plant Cell 2005, 17, 616–627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, S.W.; Li, C.H.; Cao, J.; Zhang, Y.C.; Zhang, S.Q.; Xia, Y.F.; Sun, D.Y.; Sun, Y. Altered architecture and enhanced drought tolerance in rice via the down-regulation of indole-3-acetic acid by TLD1/OsGH3.13 activation. Plant Physiol. 2009, 151, 1889–1901. [Google Scholar] [CrossRef] [Green Version]
- Du, H.; Wu, N.; Fu, J.; Wang, S.; Li, X.; Xiao, J.; Xiong, L. A GH3 family member, OsGH3-2, modulates auxin and abscisic acid levels and differentially affects drought and cold tolerance in rice. J. Exp. Bot. 2012, 63, 6467–6480. [Google Scholar] [CrossRef] [Green Version]
- Nagpal, P.; Walker, L.M.; Young, J.C.; Sonawala, A.; Timpte, C.; Estelle, M.; Reed, J.W. AXR2 Encodes a Member of the Aux/IAA Protein Family. Plant Physiol. 2000, 123, 563–574. [Google Scholar] [CrossRef] [Green Version]
- Luo, J.; Zhou, J.-J.; Zhang, J.-Z. Aux/IAA Gene Family in Plants: Molecular Structure, Regulation, and Function. Int. J. Mol. Sci. 2018, 19, 259. [Google Scholar] [CrossRef] [Green Version]
- Zenser, N.; Ellsmore, A.; Leasure, C.; Callis, J. Auxin modulates the degradation rate of Aux/IAA proteins. Proc. Natl. Acad. Sci. USA 2001, 98, 11795–11800. [Google Scholar] [CrossRef] [Green Version]
- Gray, W.M.; Kepinski, S.; Rouse, D.; Leyser, O.; Estelle, M. Auxin regulates SCFTIR1-dependent degradation of AUX/IAA proteins. Nature 2001, 414, 271–276. [Google Scholar] [CrossRef]
- Kepinski, S.; Leyser, O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nat. Cell Biol. 2005, 435, 446–451. [Google Scholar] [CrossRef]
- Tan, X.; Calderon-Villalobos, L.I.A.; Sharon, M.; Zheng, C.; Robinson, C.V.; Estelle, M.; Zheng, N. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nat. Cell Biol. 2007, 446, 640–645. [Google Scholar] [CrossRef] [PubMed]
- Maraschin Fdos, S.; Memelink, J.; Offringa, R. Auxin-induced, SCF(TIR1)-mediated poly-ubiquitination marks AUX/IAA proteins for degradation. Plant J. 2009, 59, 100–109. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, S. Molecular regulation of leaf senescence. Curr. Opin. Plant Biol. 2003, 6, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Lim, P.O.; Woo, H.R.; Gil Nam, H. Molecular genetics of leaf senescence in Arabidopsis. Trends Plant Sci. 2003, 8, 272–278. [Google Scholar] [CrossRef]
- Li, Z.; Woo, H.R.; Guo, H. Genetic redundancy of senescence-associated transcription factors in Arabidopsis. J. Exp. Bot. 2017, 69, 811–823. [Google Scholar] [CrossRef]
- Nagata, N.; Tanaka, R.; Satoh, S.; Tanaka, A. Identification of a Vinyl Reductase Gene for Chlorophyll Synthesis in Arabidopsis thaliana and Implications for the Evolution of Prochlorococcus Species. Plant Cell 2005, 17, 233–240. [Google Scholar] [CrossRef] [Green Version]
- Beale, S.I. Green genes gleaned. Trends Plant Sci. 2005, 10, 309–312. [Google Scholar] [CrossRef]
- Bollivar, D.W. Recent advances in chlorophyll biosynthesis. Photosynth. Res. 2007, 90, 173–194. [Google Scholar] [CrossRef]
- Zhang, F.; Tang, W.; Hedtke, B.; Zhong, L.; Liu, L.; Peng, L.; Lu, C.; Grimm, B.; Lin, R. Tetrapyrrole biosynthetic enzyme protoporphyrinogen IX oxidase 1 is required for plastid RNA editing. Proc. Natl. Acad. Sci. USA 2014, 111, 2023–2028. [Google Scholar] [CrossRef] [Green Version]
- Walker, J.C.; Willows, R. Mechanism and regulation of Mg-chelatase. Biochem. J. 1997, 327, 321–333. [Google Scholar] [CrossRef] [Green Version]
- Masuda, T. Recent overview of the Mg branch of the tetrapyrrole biosynthesis leading to chlorophylls. Photosynth. Res. 2008, 96, 121–143. [Google Scholar] [CrossRef] [PubMed]
- Papenbrock, J.; Grafe, S.; Kruse, E.; Hanel, F.; Grimm, B. Mg-chelatase of tobacco: Identification of a Chl D cDNA sequence encoding a third subunit, analysis of the interaction of the three subunits with the yeast two-hybrid system, and reconstitution of the enzyme activity by co-expression of recombinant CHL D, CHL H and CHL I. Plant J. 1997, 12, 981–990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sawicki, A.; Zhou, S.; Kwiatkowski, K.; Luo, M.; Willows, R. 1-N-histidine phosphorylation of ChlD by the AAA+ ChlI2 stimulates magnesium chelatase activity in chlorophyll synthesis. Biochem. J. 2017, 474, 2095–2105. [Google Scholar] [CrossRef] [PubMed]
- Farmer, D.A.; Brindley, A.A.; Hitchcock, A.; Jackson, P.J.; Johnson, B.; Dickman, M.J.; Hunter, C.N.; Reid, J.D.; Adams, N.B.P. The ChlD subunit links the motor and porphyrin binding subunits of magnesium chelatase. Biochem. J. 2019, 476, 1875–1887. [Google Scholar] [CrossRef] [Green Version]
- Ikegami, A.; Yoshimura, N.; Motohashi, K.; Takahashi, S.; Romano, P.G.N.; Hisabori, T.; Takamiya, K.-I.; Masuda, T. The CHLI1 Subunit ofArabidopsis thalianaMagnesium Chelatase Is a Target Protein of the Chloroplast Thioredoxin. J. Biol. Chem. 2007, 282, 19282–19291. [Google Scholar] [CrossRef] [Green Version]
- Kobayashi, K.; Mochizuki, N.; Yoshimura, N.; Motohashi, K.; Hisabori, T.; Masuda, T. Functional analysis of Arabidopsis thaliana isoforms of the Mg-chelatase CHLI subunit. Photochem. Photobiol. Sci. 2008, 7, 1188–1195. [Google Scholar] [CrossRef]
- Block, M.A.; Tewari, A.K.; Albrieux, C.; Maréchal, E.; Joyard, J. The plant S -adenosyl-l -methionine: Mg-protoporphyrin IX methyltransferase is located in both envelope and thylakoid chloroplast membranes. JBIC J. Biol. Inorg. Chem. 2002, 269, 240–248. [Google Scholar] [CrossRef]
- Alawady, A.; Reski, R.; Yaronskaya, E.; Grimm, B. Cloning and expression of the tobacco CHLM sequence encoding Mg protoporphyrin IX methyltransferase and its interaction with Mg chelatase. Plant Mol. Biol. 2005, 57, 679–691. [Google Scholar] [CrossRef]
- Alboresi, A.; Ballottari, M.; Hienerwadel, R.; Giacometti, G.M.; Morosinotto, T. Antenna complexes protect Photosystem I from Photoinhibition. BMC Plant Biol. 2009, 9, 71. [Google Scholar] [CrossRef] [Green Version]
- Fox, K.F.; Ünlü, C.; Balevičius, V.; Ramdour, B.N.; Kern, C.; Pan, X.; Li, M.; Van Amerongen, H.; Duffy, C.D.P. A possible molecular basis for photoprotection in the minor antenna proteins of plants. Biochim. Biophys. Acta (BBA) Bioenerg. 2018, 1859, 471–481. [Google Scholar] [CrossRef]
- De Bianchi, S.; Betterle, N.; Kouril, R.; Cazzaniga, S.; Boekema, E.; Bassi, R.; Osto, L.D. Arabidopsis Mutants Deleted in the Light-Harvesting Protein Lhcb4 Have a Disrupted Photosystem II Macrostructure and Are Defective in Photoprotection. Plant Cell 2011, 23, 2659–2679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adamiec, M.; Gibasiewicz, K.; Luciński, R.; Giera, W.; Chełminiak, P.; Szewczyk, S.; Sipińska, W.; Van Grondelle, R.; Jackowski, G. Excitation energy transfer and charge separation are affected in Arabidopsis thaliana mutants lacking light-harvesting chlorophyll a/b binding protein Lhcb3. J. Photochem. Photobiol. B Biol. 2015, 153, 423–428. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-E.; Ma, J.; Wu, N.; Su, Y.-Q.; Zhang, Z.-W.; Yuan, M.; Zhang, H.-Y.; Zeng, X.-Y.; Yuan, S. The roles of Arabidopsis proteins of Lhcb4, Lhcb5 and Lhcb6 in oxidative stress under natural light conditions. Plant Physiol. Biochem. 2018, 130, 267–276. [Google Scholar] [CrossRef] [PubMed]
- Ihalainen, J.A.; Klimmek, F.; Ganeteg, U.; Van Stokkum, I.H.; Van Grondelle, R.; Jansson, S.; Dekker, J.P. Excitation energy trapping in photosystem I complexes depleted in Lhca1 and Lhca4. FEBS Lett. 2005, 579, 4787–4791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Otani, T.; Yamamoto, H.; Shikanai, T. Stromal Loop of Lhca6 is Responsible for the Linker Function Required for the NDH–PSI Supercomplex Formation. Plant Cell Physiol. 2017, 58, 851–861. [Google Scholar] [CrossRef] [Green Version]
- Wright, I.J.; Reich, P.B.; Westoby, M.; Ackerly, D.; Baruch, Z.; Bongers, F.; Cavender-Bares, J.; Chapin, T.; Cornelissen, J.H.C.; Diemer, M.; et al. The worldwide leaf economics spectrum. Nature 2004, 428, 821–827. [Google Scholar] [CrossRef]
- Delvallé, D.; Dumez, S.; Wattebled, F.; Roldán, I.; Planchot, V.; Berbezy, P.; Colonna, P.; Vyas, D.; Chatterjee, M.; Ball, S.; et al. Soluble starch synthase I: A major determinant for the synthesis of amylopectin in Arabidopsis thaliana leaves. Plant J. 2005, 43, 398–412. [Google Scholar] [CrossRef]
- Szydlowski, N.; Ragel, P.; Raynaud, S.; Lucas, M.M.; Roldán, I.; Montero, M.; Muñoz, F.J.; Ovecka, M.; Bahaji, A.; Planchot, V.; et al. Starch Granule Initiation in Arabidopsis Requires the Presence of Either Class IV or Class III Starch Synthases. Plant Cell 2009, 21, 2443–2457. [Google Scholar] [CrossRef] [Green Version]
- Crumpton-Taylor, M.; Pike, M.; Lu, K.; Hylton, C.M.; Feil, R.; Eicke, S.; Lunn, J.E.; Zeeman, S.C.; Smith, A.M. Starch synthase 4 is essential for coordination of starch granule formation with chloroplast division during Arabidopsis leaf expansion. New Phytol. 2013, 200, 1064–1075. [Google Scholar] [CrossRef]
- Maloney, V.; Park, J.-Y.; Unda, F.; Mansfield, S.D. Sucrose phosphate synthase and sucrose phosphate phosphatase interact in planta and promote plant growth and biomass accumulation. J. Exp. Bot. 2015, 66, 4383–4394. [Google Scholar] [CrossRef] [Green Version]
- Matile, P.; Hörtensteiner, S.; Thomas, H. CHLOROPHYLL DEGRADATION. Annu. Rev. Plant Biol. 1999, 50, 67–95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, S.-Y.; Yu, J.-W.; Park, J.-S.; Li, J.; Yoo, S.-C.; Lee, N.-Y.; Lee, S.-K.; Jeong, S.-W.; Seo, H.S.; Koh, H.-J.; et al. The Senescence-Induced Staygreen Protein Regulates Chlorophyll Degradation. Plant Cell 2007, 19, 1649–1664. [Google Scholar] [CrossRef] [PubMed] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Ren, Y.; Kang, X. Study on Gene Differential Expression in Tetraploid Populus Leaves. Forests 2020, 11, 1233. https://doi.org/10.3390/f11111233
Zhang Y, Ren Y, Kang X. Study on Gene Differential Expression in Tetraploid Populus Leaves. Forests. 2020; 11(11):1233. https://doi.org/10.3390/f11111233
Chicago/Turabian StyleZhang, Ying, Yongyu Ren, and Xiangyang Kang. 2020. "Study on Gene Differential Expression in Tetraploid Populus Leaves" Forests 11, no. 11: 1233. https://doi.org/10.3390/f11111233




