Mycorrhizal Fungi Synergistically Promote the Growth and Secondary Metabolism of Cyclocarya paliurus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material and Experimental Design
2.2. Observation on Colonization of Mycorrhizal Fungi
2.3. Measurements of Plant Growth, Biomass, Root Traits and Photosynthetic Rate
2.4. Measurement of Phytohormones
2.5. Measurement of Mineral Nutrient
2.6. Extraction and Determination of Triterpenoids and Flavonoids
2.7. RNA Extraction, Sequencing, and Differential Expression Analysis
2.8. Identification of Fungal Microbiota Diversity in Roots
2.9. Data Analysis
3. Results and Discussion
3.1. Variation in Colonization Rate
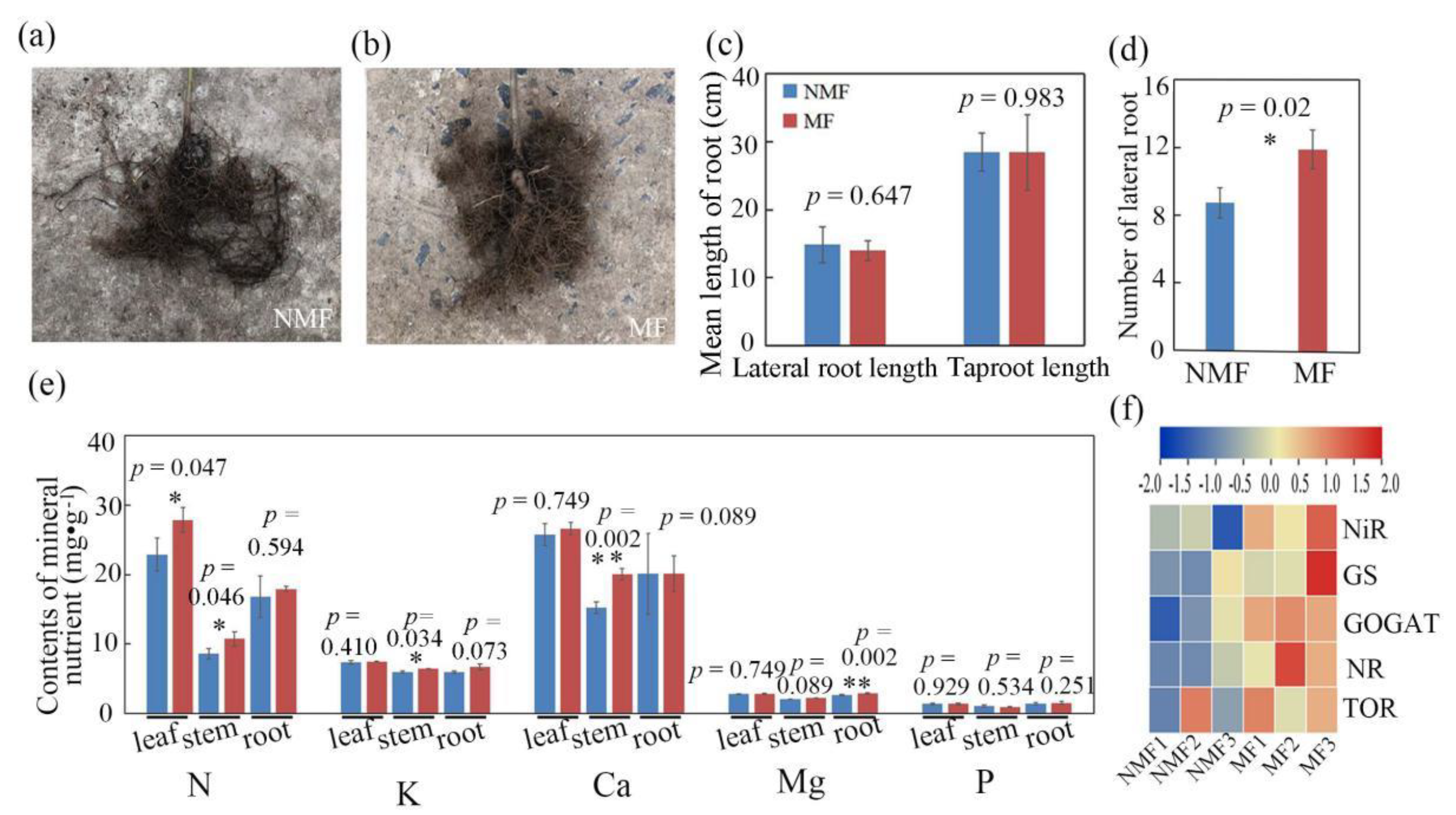
3.2. Variation in Growth and Biomass Accumulation
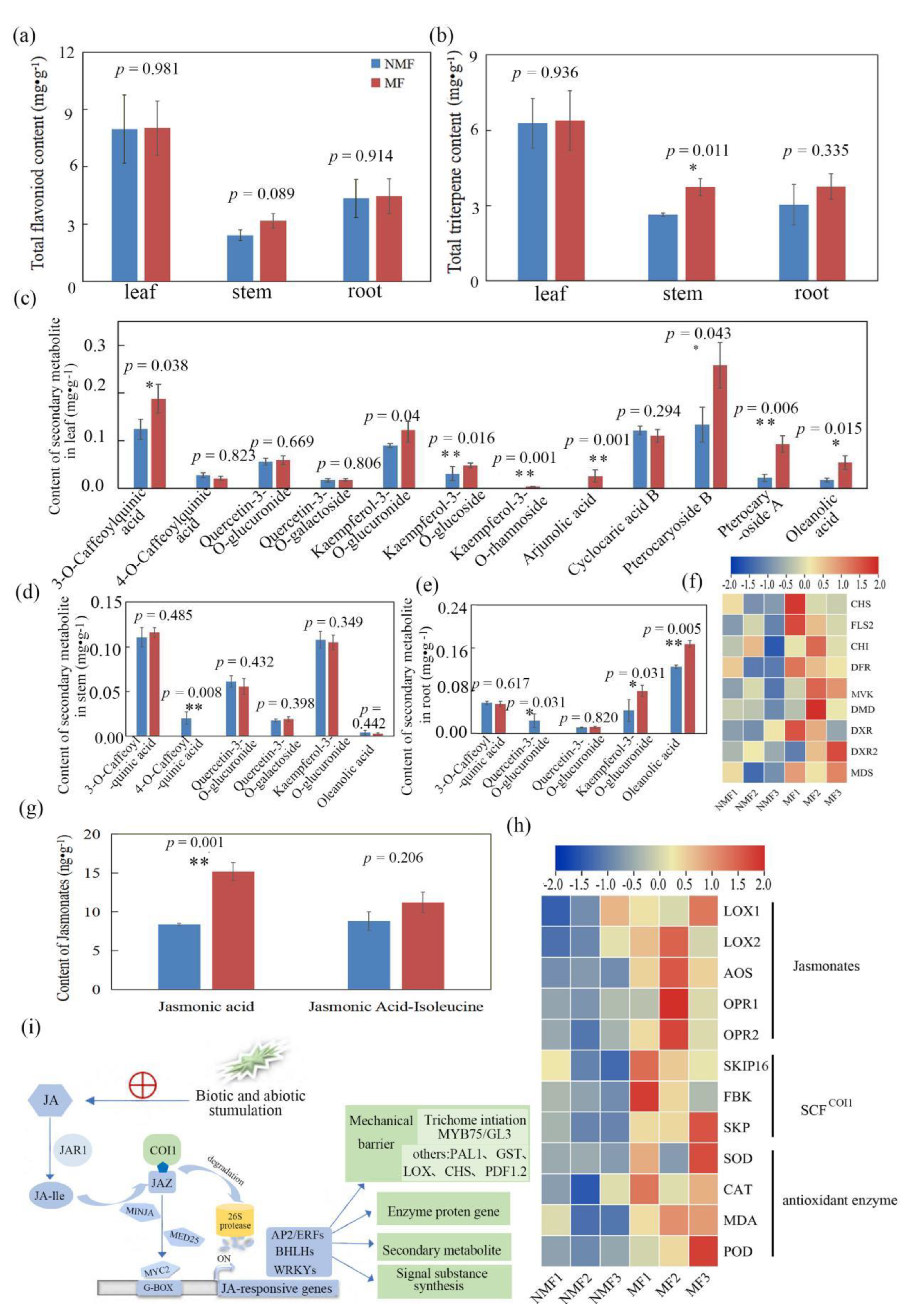
3.3. Variation in Secondary Metabolism within Plants
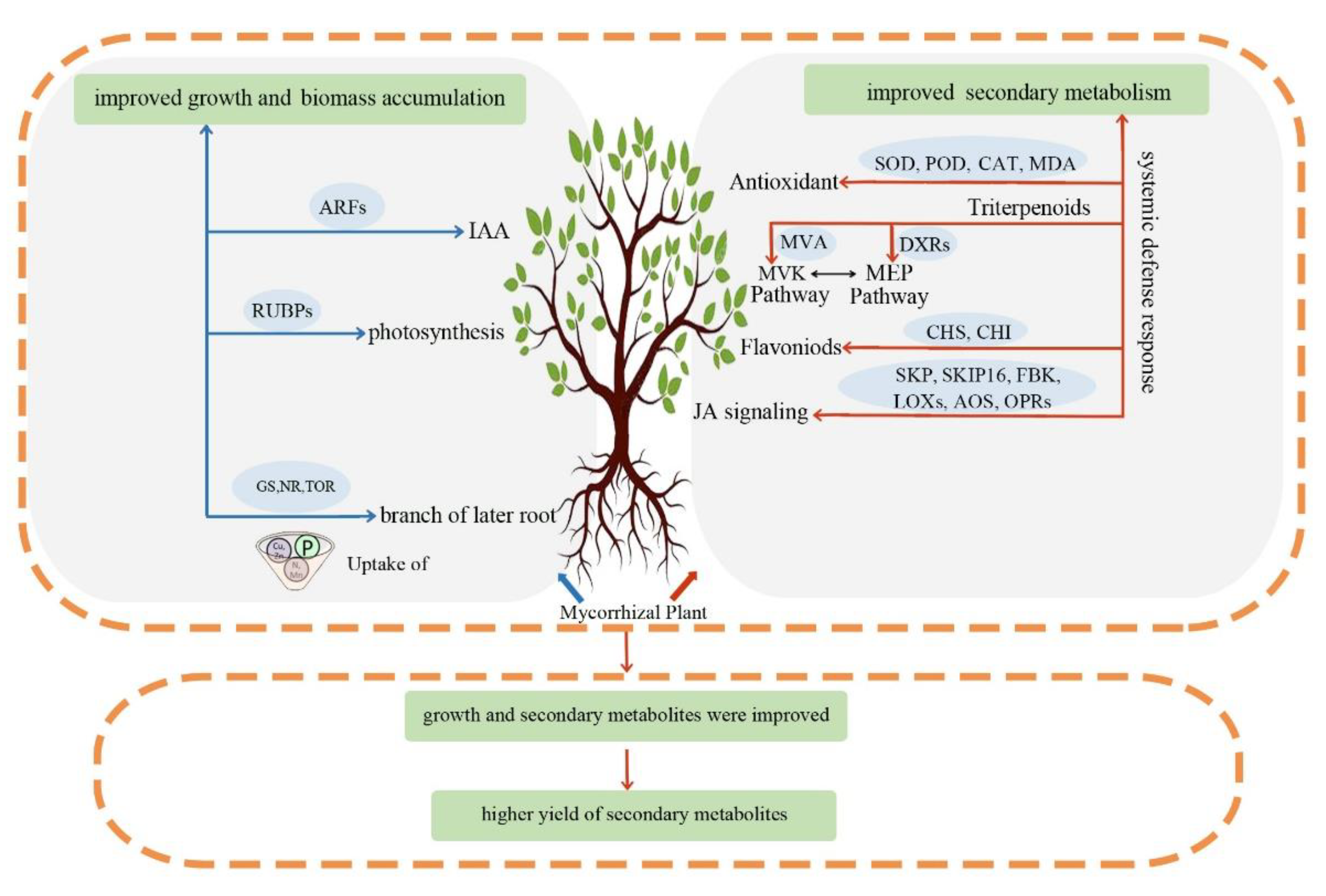
3.4. Variation in Yield of Secondary Metabolites per Plants
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Fang, S.; Yang, W.; Chu, X.; Shang, X.; She, C.; Fu, X. Provenance and temporal variations in selected flavonoids in leaves of Cyclocarya paliurus. Food Chem. 2011, 124, 1382–1386. [Google Scholar] [CrossRef]
- Lin, Z.; Wu, Z.-F.; Jiang, C.-H.; Zhang, Q.-W.; Ouyang, S.; Che, C.-T.; Zhang, J.; Yin, Z.-Q. The chloroform extract of Cyclocarya paliurus attenuates high-fat diet induced non-alcoholic hepatic steatosis in Sprague Dawley rats. Phytomedicine 2016, 23, 1475–1483. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Jiang, C.; Fang, S.; Wang, J.; Ji, Y.; Shang, X.; Ni, Y.; Yin, Z.; Zhang, J. Antihyperglycemic, antihyperlipidemic and antioxidant effects of ethanol and aqueous extracts of Cyclocarya paliurus leaves in type 2 diabetic rats. J. Ethnopharmacol. 2013, 150, 1119–1127. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.F.; Gao, T.H.; Zhong, R.L.; Lin, Z.; Jiang, C.H.; Ouyang, S.; Zhao, M.; Che, C.T.; Zhang, J.; Yin, Z.Q. Antihyperlipidaemic effect of triterpenic acid-enriched fraction from Cyclocarya paliurus leaves in hyperlipidaemic rats. Pharm. Boil. 2017, 55, 712–721. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, B.; Shang, X.; Fang, S.; Li, Q.; Fu, X.; Su, J. Integrated Effects of Light Intensity and Fertilization on Growth and Flavonoid Accumulation in Cyclocarya paliurus. J. Agric. Food Chem. 2012, 60, 6286–6292. [Google Scholar] [CrossRef] [PubMed]
- Deng, B.; Li, Y.; Lei, G.; Liu, G. Effects of nitrogen availability on mineral nutrient balance and flavonoid accumulation in Cyclocarya paliurus. Plant Physiol. Biochem. 2019, 135, 111–118. [Google Scholar] [CrossRef]
- Deng, B.; Li, Y.; Xu, D.; Ye, Q.; Liu, G. Nitrogen availability alters flavonoid accumulation in Cyclocarya paliurus via the effects on the internal carbon/nitrogen balance. Sci. Rep. 2019, 9, 2370. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Zhang, M.; Zhao, T.; Deng, B. Phosphate availability regulates flavonoid accumulation associated with photosynthetic carbon partitioning in Cyclocarya paliurus. Physiol. Plant. 2021, 173, 1956–1966. [Google Scholar] [CrossRef]
- Brundrett, M.C.; Tedersoo, L. Evolutionary history of mycorrhizal symbioses and global host plant diversity. New Phytol. 2018, 220, 1108–1115. [Google Scholar] [CrossRef] [Green Version]
- Mandal, S.; Upadhyay, S.; Singh, V.P.; Kapoor, R. Enhanced production of steviol glycosides in mycorrhizal plants: A concerted effect of arbuscular mycorrhizal symbiosis on transcription of biosynthetic genes. Plant Physiol. Biochem. 2015, 89, 100–106. [Google Scholar] [CrossRef]
- Mandal, S.; Upadhyay, S.; Wajid, S.; Ram, M.; Jain, D.C.; Singh, V.P.; Abdin, M.Z.; Kapoor, R. Arbuscular mycorrhiza increase artemisinin accumulation in Artemisia annua by higher expression of key biosynthesis genes via enhanced jasmonic acid levels. Mycorrhiza 2015, 25, 345–357. [Google Scholar] [CrossRef]
- Sheteiwy, M.S.; Elgawad, H.A.; Xiong, Y.; Macovei, A.; Brestic, M.; Skalicky, M.; Shaghaleh, H.; Hamoud, Y.A.; El-Sawah, A.M. Inoculation with Bacillus amyloliquefaciens and mycorrhiza confers tolerance to drought stress and improve seed yield and quality of soybean plant. Physiol. Plant. 2021, 172, 2153–2169. [Google Scholar] [CrossRef] [PubMed]
- Nagy, L.G.; Riley, R.; Tritt, A.; Adam, C.; Daum, C.; Floudas, D.; Sun, H.; Yadav, J.S.; Pangilinan, J.; Larsson, K.-H.; et al. Comparative Genomics of Early-Diverging Mushroom-Forming Fungi Provides Insights into the Origins of Lignocellulose Decay Capabilities. Mol. Biol. Evol. 2016, 33, 959–970. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plett, J.M.; Daguerre, Y.; Wittulsky, S.; Vayssières, A.; Deveau, A.; Melton, S.J.; Kohler, A.; Morrell-Falvey, J.L.; Brun, A.; Veneault-Fourrey, C.; et al. Effector MiSSP7 of the mutualistic fungus Laccaria bicolor stabilizes the Populus JAZ6 protein and represses jasmonic acid (JA) responsive genes. Proc. Natl. Acad. Sci. USA 2014, 111, 8299–8304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, J.-H.; Halitschke, R.; Kim, H.B.; Baldwin, I.T.; Feldmann, K.A.; Feyereisen, R. A knock-out mutation in allene oxide synthase results in male sterility and defective wound signal transduction in Arabidopsis due to a block in jasmonic acid biosynthesis. Plant J. 2002, 31, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Gutjahr, C. Phytohormone signaling in arbuscular mycorrhiza development. Curr. Opin. Plant Biol. 2014, 20, 26–34. [Google Scholar] [CrossRef] [PubMed]
- Kapoor, R.; Anand, G.; Gupta, P.; Mandal, S. Insight into the mechanisms of enhanced production of valuable terpenoids by arbuscular mycorrhiza. Phytochem. Rev. 2017, 16, 677–692. [Google Scholar] [CrossRef]
- Le Roy, J.; Huss, B.; Creach, A.; Hawkins, S.; Neutelings, G. Glycosylation is a major regulator of phenylpropanoid availability and biological activity in plants. Front. Plant Sci. 2016, 7, 735. [Google Scholar] [CrossRef] [Green Version]
- Augustin, J.M.; Drok, S.; Shinoda, T.; Sanmiya, K.; Nielsen, J.K.; Khakimov, B.; Olsen, C.E.; Hansen, E.H.; Kuzina, V.; Ekstrøm, C.T.; et al. UDP-Glycosyltransferases from the UGT73C Subfamily in Barbarea vulgaris Catalyze Sapogenin 3-O-Glucosylation in Saponin-Mediated Insect Resistance. Plant Physiol. 2012, 160, 1881–1895. [Google Scholar] [CrossRef] [Green Version]
- Nielsen, J.K.; Nagao, T.; Okabe, H.; Shinoda, T. Resistance in the Plant, Barbarea vulgaris, and Counter-Adaptations in Flea Beetles Mediated by Saponins. J. Chem. Ecol. 2010, 36, 277–285. [Google Scholar] [CrossRef]
- Fang, S.; Wang, J.; Wei, Z.; Zhu, Z. Methods to break seed dormancy in Cyclocarya paliurus (Batal)Iljinskaja. Sci. Hortic. 2006, 110, 305–309. [Google Scholar] [CrossRef]
- Zhang, Z.F.; Zhang, J.C.; Zhou, L.W.; Xu, G.P.; Li, Y.Q. Effects of arbuscular mycorrhizal fungi on the growth of afforestation seedlings in a rocky desertification area. Chin. J. Ecol. 2018, 37, 2917–2934. (In Chinese) [Google Scholar]
- Massenssini, A.M.; Bonduki, V.H.A.; Tótola, M.R.; Ferreira, F.A.; Costa, M.D. Arbuscular mycorrhizal associations and occurrence of dark septate endophytes in the roots of Brazilian weed plants. Mycorrhiza 2014, 24, 153–159. [Google Scholar] [CrossRef] [PubMed]
- Bao, J.; Cai, Y.; Sun, M.; Wang, G.; Corke, H. Anthocyanins, flavonols, and free radical scavenging activity of Chinese bayberry (Myrica rubra) extracts and their color properties and stability. J. Agric. Food Chem. 2005, 53, 2327–2332. [Google Scholar] [CrossRef]
- Fan, J.-P.; He, C.-H. Simultaneous quantification of three major bioactive triterpene acids in the leaves of Diospyros kaki by high-performance liquid chromatography method. J. Pharm. Biomed. Anal. 2006, 41, 950–956. [Google Scholar] [CrossRef]
- Cao, Y.; Fang, S.; Yin, Z.; Fu, X.; Shang, X.; Yang, W.; Yang, H. Chemical fingerprint and multicomponent quantitative analysis for the quality evaluation of Cyclocarya paliurus leaves by HPLC–Q–TOF–MS. Molecules 2017, 22, 1927. [Google Scholar] [CrossRef] [Green Version]
- Wu, T.G.; Mitchell, B.M.; Carothers, T.S.; Coats, D.K.; Brady-McCreery, K.M.; Paysse, E.A.; Wilhelmus, K.R. Molecular analysis of the pediatric ocular surface for fungi. Curr. Eye Res. 2003, 26, 33–36. [Google Scholar] [CrossRef]
- Mao, J.; Li, R.; Jing, Y.; Ning, D.; Li, Y.; Chen, H. Arbuscular mycorrhizal fungi associated with walnut trees and their effects on seedling growth. J. For. Environ. 2022, 42, 71–80. (In Chinese) [Google Scholar]
- Mandal, S.; Evelin, H.; Giri, B.; Singh, V.P.; Kapoor, R. Arbuscular mycorrhiza enhances the production of stevioside and rebaudioside-A in Stevia rebaudiana via nutritional and non-nutritional mechanisms. Appl. Soil Ecol. 2013, 72, 187–194. [Google Scholar] [CrossRef]
- Rasouli-Sadaghiani, M.; Hassani, A.; Barin, M.; Danesh, Y.R.; Sefidkon, F. Effects of arbuscular mycorrhizal (AM) fungi on growth, essential oil production and nutrients uptake in basil. J. Med. Plant. Res. 2010, 4, 2222–2228. [Google Scholar]
- Fitze, D.; Wiepning, A.; Kaldorf, M.; Ludwig-Müller, J. Auxins in the development of an arbuscular mycorrhizal symbiosis in maize. J. Plant Physiol. 2005, 162, 1210–1219. [Google Scholar] [CrossRef] [PubMed]
- Hanlon, M.; Coenen, C. Genetic evidence for auxin involvement in arbuscular mycorrhiza initiation. New Phytol. 2011, 189, 701–709. [Google Scholar] [CrossRef] [PubMed]
- Liao, D.; Chen, X.; Chen, A.; Wang, H.; Liu, J.; Liu, J.; Gu, M.; Sun, S.; Xu, G. The Characterization of Six Auxin-Induced Tomato GH3 Genes Uncovers a Member, SlGH3.4, Strongly Responsive to Arbuscular Mycorrhizal Symbiosis. Plant Cell Physiol. 2015, 56, 674–687. [Google Scholar] [CrossRef] [PubMed]
- Guilfoyle, T.J.; Hagen, G. Auxin response factors. Curr. Opin. Plant Biol. 2007, 10, 453–460. [Google Scholar] [CrossRef]
- Chen, J.; Wang, P.; Mi, H.-L.; Chen, G.-Y.; Xu, D.-Q. Reversible association of ribulose-1, 5-bisphosphate carboxylase/oxygenase activase with the thylakoid membrane depends upon the ATP level and pH in rice without heat stress. J. Exp. Bot. 2010, 61, 2939–2950. [Google Scholar] [CrossRef] [Green Version]
- Genre, A.; Lanfranco, L.; Perotto, S.; Bonfante, P. Unique and common traits in mycorrhizal symbioses. Nat. Rev. Microbiol. 2020, 18, 649–660. [Google Scholar] [CrossRef]
- Dong, N.Q.; Lin, H.X. Contribution of phenylpropanoid metabolism to plant development and plant–environment interactions. J. Integr. Plant Biol. 2021, 63, 180–209. [Google Scholar] [CrossRef]
- Lin, J.-S.; Huang, X.-X.; Li, Q.; Cao, Y.; Bao, Y.; Meng, X.-F.; Li, Y.-J.; Fu, C.; Hou, B.-K. UDP-glycosyltransferase 72B1 catalyzes the glucose conjugation of monolignols and is essential for the normal cell wall lignification in Arabidopsis thaliana. Plant J. 2016, 88, 26–42. [Google Scholar] [CrossRef] [Green Version]
- Wu, Q.-S.; Zou, Y.-N.; He, X.-H. Contributions of arbuscular mycorrhizal fungi to growth, photosynthesis, root morphology and ionic balance of citrus seedlings under salt stress. Acta Physiol. Plant. 2010, 32, 297–304. [Google Scholar] [CrossRef]
- Alaux, P.L.; Naveau, F.; Declerck, S.; Cranenbrouck, S. Common mycorrhizal network induced JA/ET genes expression in healthy potato plants connected to potato plants y by Phytophthora infestans. Front. Plant Sci. 2020, 11, 602. [Google Scholar] [CrossRef]
- Oliveira, R.S.; Franco, A.R.; Vosatka, M.; Castro, P.M.L. Management of nursery practices for efficient ectomycorrhizal fungi application in the production of Quercus ilex. Symbiosis 2010, 52, 125–131. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.; Shi, L.; Li, L.; Fu, L.; Liu, Y.; Xiong, Y.; Sheen, J. Integration of nutrient, energy, light, and hormone signaling via TOR in plants. J. Exp. Bot. 2019, 70, 2227–2238. [Google Scholar] [CrossRef] [PubMed]
- Crepin, N.; Rolland, F. SnRK1 activation, signaling, and networking for energy homeostasis. Curr. Opin. Plant Biol. 2019, 51, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Shirley, B.W.; Kubasek, W.L.; Storz, G.; Bruggemann, E.; Koornneef, M.; Ausubel, F.M.; Goodman, H.M. Analysis of Arabidopsis mutants deficient in flavonoid biosynthesis. Plant J. 1995, 8, 659–671. [Google Scholar] [CrossRef] [PubMed]
- Winkel-Shirley, B. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 2001, 126, 485–493. [Google Scholar] [CrossRef] [Green Version]
- Lv, Z.Y.; Zhang, F.Y.; Pan, Q.F.; Fu, X.Q.; Jiang, W.M.; Shen, Q.; Yan, T.X.; Shi, P.; Lu, X.; Sun, X.F.; et al. Branch pathway blocking in Artemisia annua is a useful method for obtaining high yield artemisnin. Plant Cell Physiol. 2016, 57, 588–602. [Google Scholar] [CrossRef] [Green Version]
- Cui, L.G.; Shan, J.X.; Shi, M.; Gao, J.P.; Lin, H.X. The miR156-SPL 9-DFR pathway coordinates the relationship between development and abiotic stress tolerance in plants. Plant J. 2014, 80, 1108–1117. [Google Scholar] [CrossRef]
- Li, Y.; Liu, Z.; Hou, H.; Lei, H.; Zhu, X.; Li, X.; He, X.; Tian, C. Arbuscular mycorrhizal fungi-enhanced resistance against Phytophthora sojae infection on soybean leaves is mediated by a network involving hydrogen peroxide, jasmonic acid, and the metabolism of carbon and nitrogen. Acta Physiol. Plant. 2013, 35, 3465–3475. [Google Scholar] [CrossRef]
- Jung, S.C.; Martinez-Medina, A.; Lopez-Raez, J.A.; Pozo, M.J. Mycorrhiza-Induced Resistance and Priming of Plant Defenses. J. Chem. Ecol. 2012, 38, 651–664. [Google Scholar] [CrossRef]
- Plett, J.M.; Khachane, A.; Ouassou, M.; Sundberg, B.; Kohler, A.; Martin, F. Ethylene and jasmonic acid act as negative modulators during mutualistic symbiosis between Laccaria bicolor and Populus roots. New Phytol. 2014, 202, 270–286. [Google Scholar] [CrossRef]
- Fonseca, S.; Chico, J.M.; Solano, R. The jasmonate pathway: The ligand, the receptor and the core signaling module. Curr. Opin. Plant Biol. 2009, 12, 539–547. [Google Scholar] [CrossRef] [PubMed]
- Katsir, L.; Chung, H.S.; Koo, A.J.; Howe, G.A. Jasmonate signaling: A conserved mechanism of hormone sensing. Curr. Opin. Plant Biol. 2008, 11, 428–435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stamp, N. Out of the Quagmire of Plant Defense Hypotheses. Q. Rev. Biol. 2003, 78, 23–55. [Google Scholar] [CrossRef] [PubMed]
- Larbat, R.; Robin, C.; Lillo, C.; Drengstig, T.; Ruoff, P. Modeling the diversion of primary carbon flux into secondary metabolism under variable nitrate and light/dark conditions. J. Theor. Biol. 2016, 402, 144–157. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, T.; Yu, B.; Zhang, M.; Chen, S.; Deng, B. Mycorrhizal Fungi Synergistically Promote the Growth and Secondary Metabolism of Cyclocarya paliurus. Forests 2022, 13, 2188. https://doi.org/10.3390/f13122188
Zhao T, Yu B, Zhang M, Chen S, Deng B. Mycorrhizal Fungi Synergistically Promote the Growth and Secondary Metabolism of Cyclocarya paliurus. Forests. 2022; 13(12):2188. https://doi.org/10.3390/f13122188
Chicago/Turabian StyleZhao, Tingting, Bangyou Yu, Mengjia Zhang, Shuying Chen, and Bo Deng. 2022. "Mycorrhizal Fungi Synergistically Promote the Growth and Secondary Metabolism of Cyclocarya paliurus" Forests 13, no. 12: 2188. https://doi.org/10.3390/f13122188



