Population Genetics and Development of a Core Collection from Elite Germplasms of Xanthoceras sorbifolium Based on Genome-Wide SNPs
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. DNA Extraction and Genotyping
2.3. Phylogenetic and Population Structure Analysis
2.4. Genetic Diversity Analysis
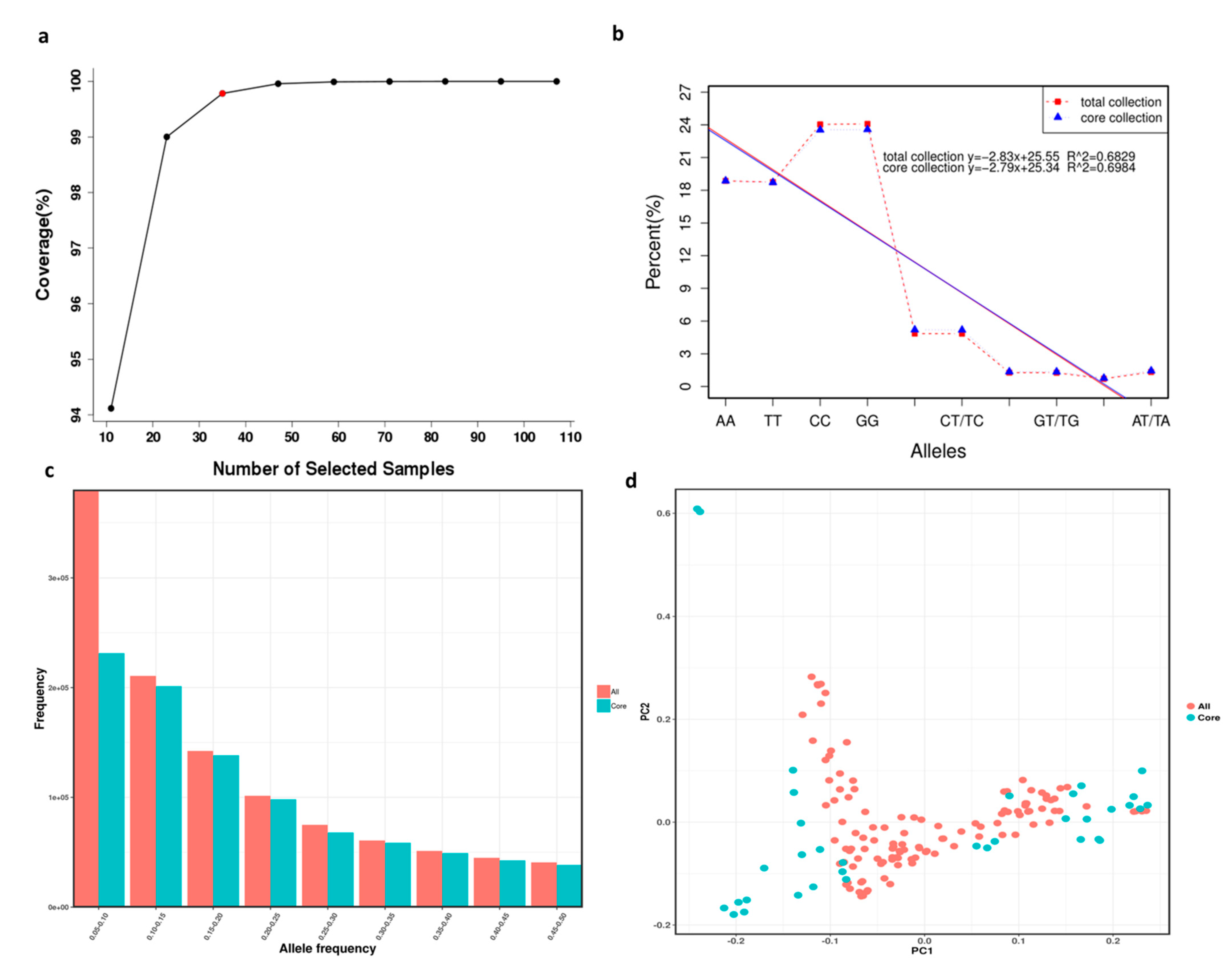
2.5. Core Collection Establishment and Evaluation
3. Results
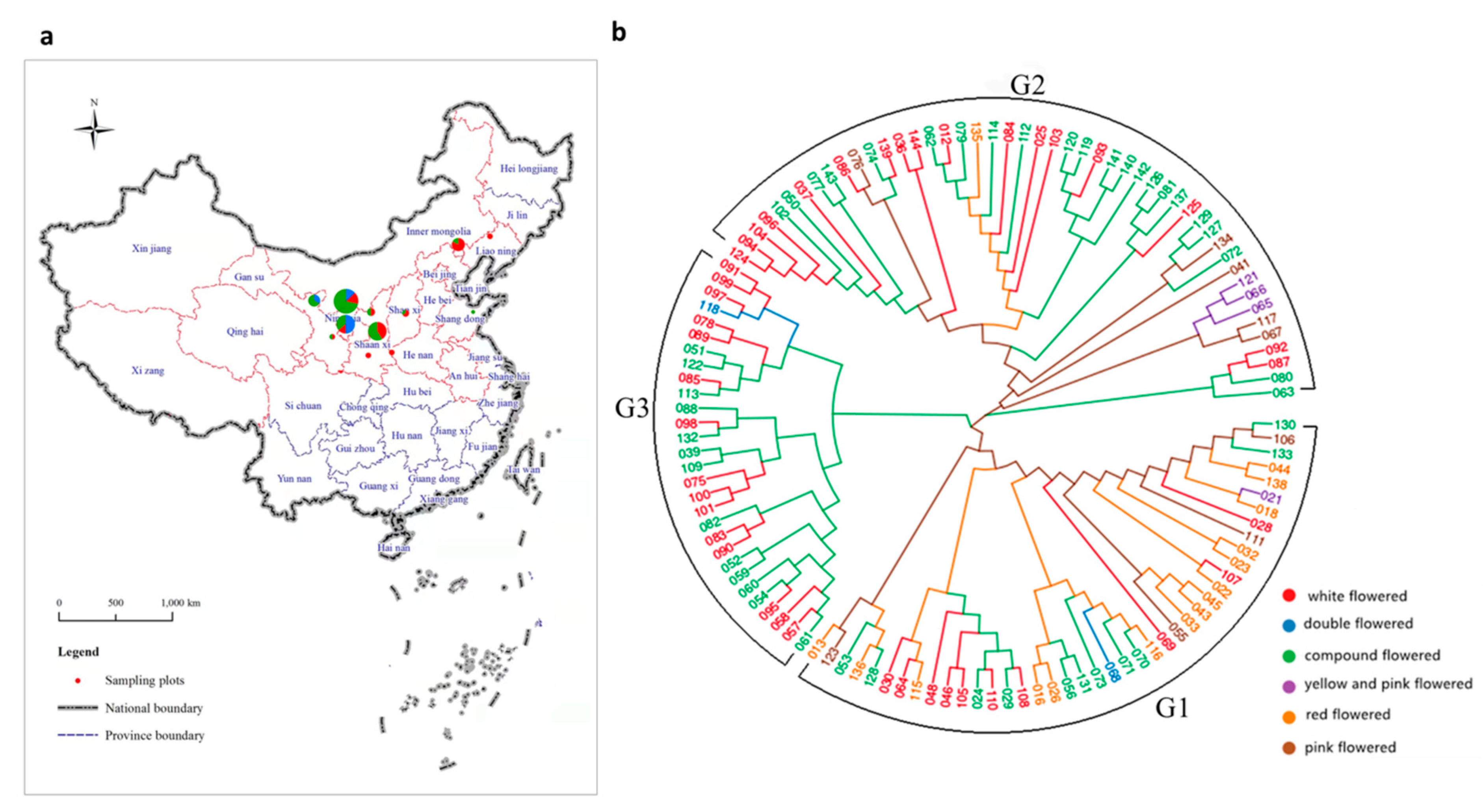
3.1. Sequencing of X. sorbifolium among 119 Elite Germplasms
3.2. Phylogenetic Analysis
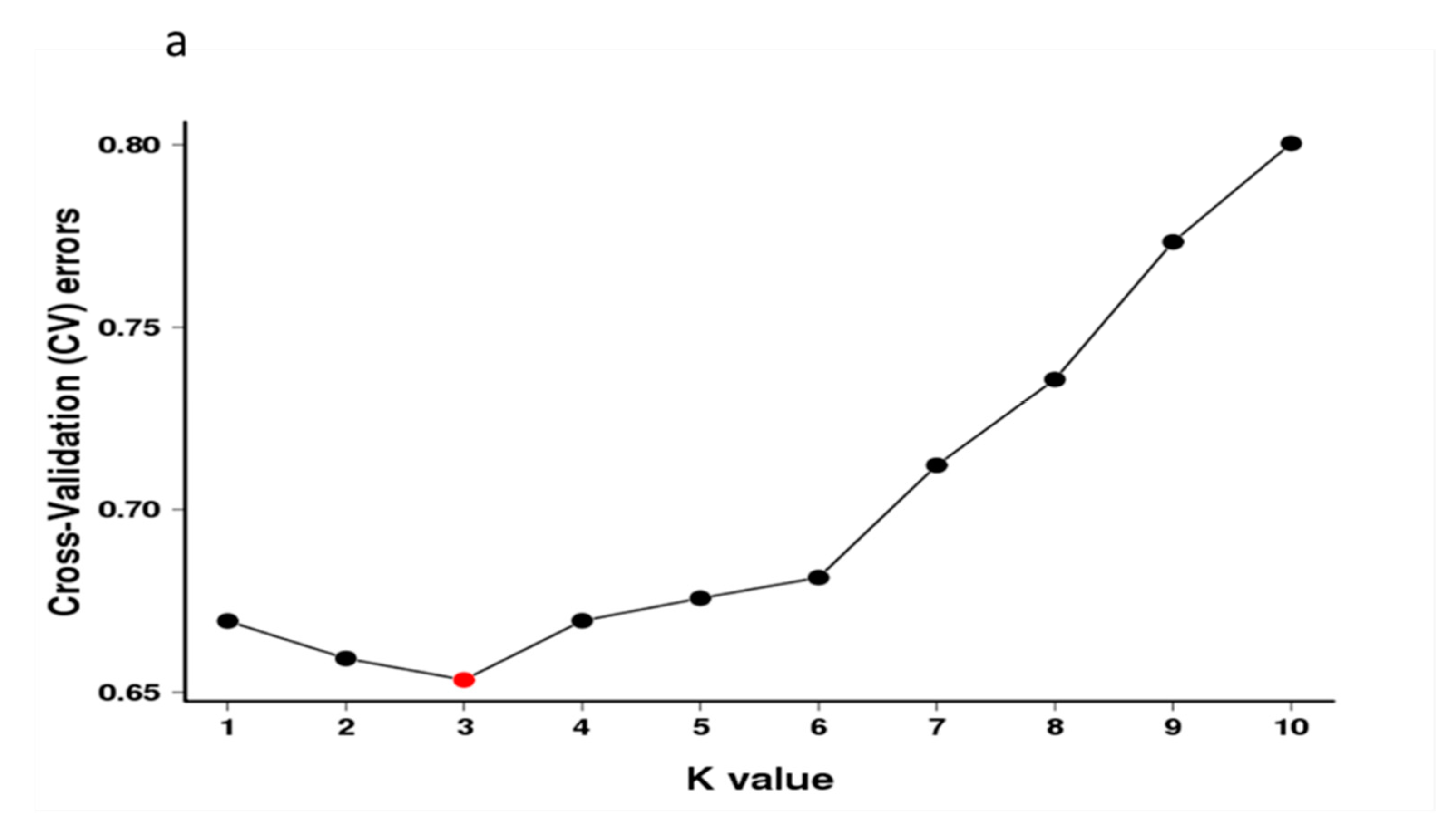
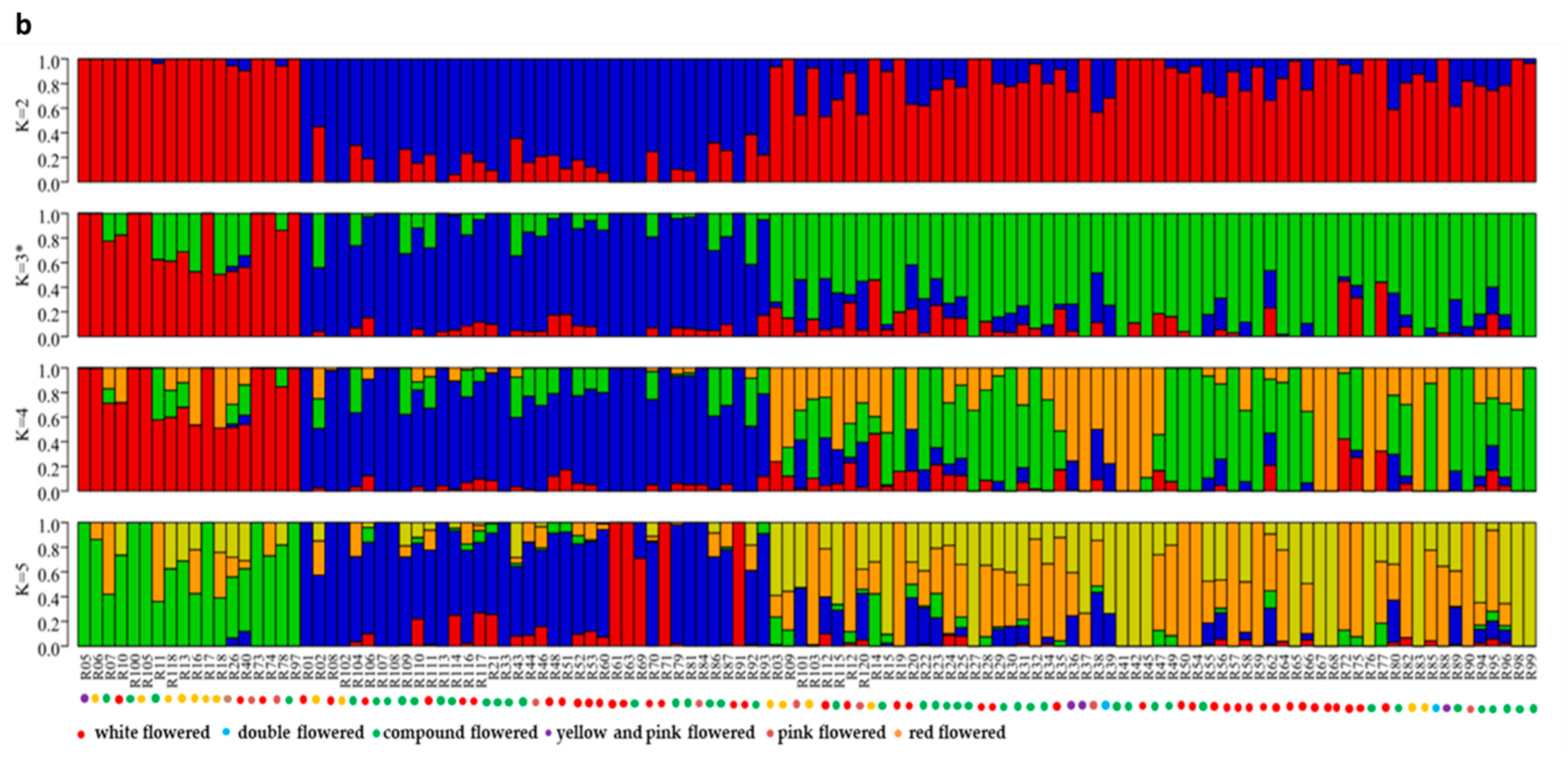
3.3. Population Structure Analysis
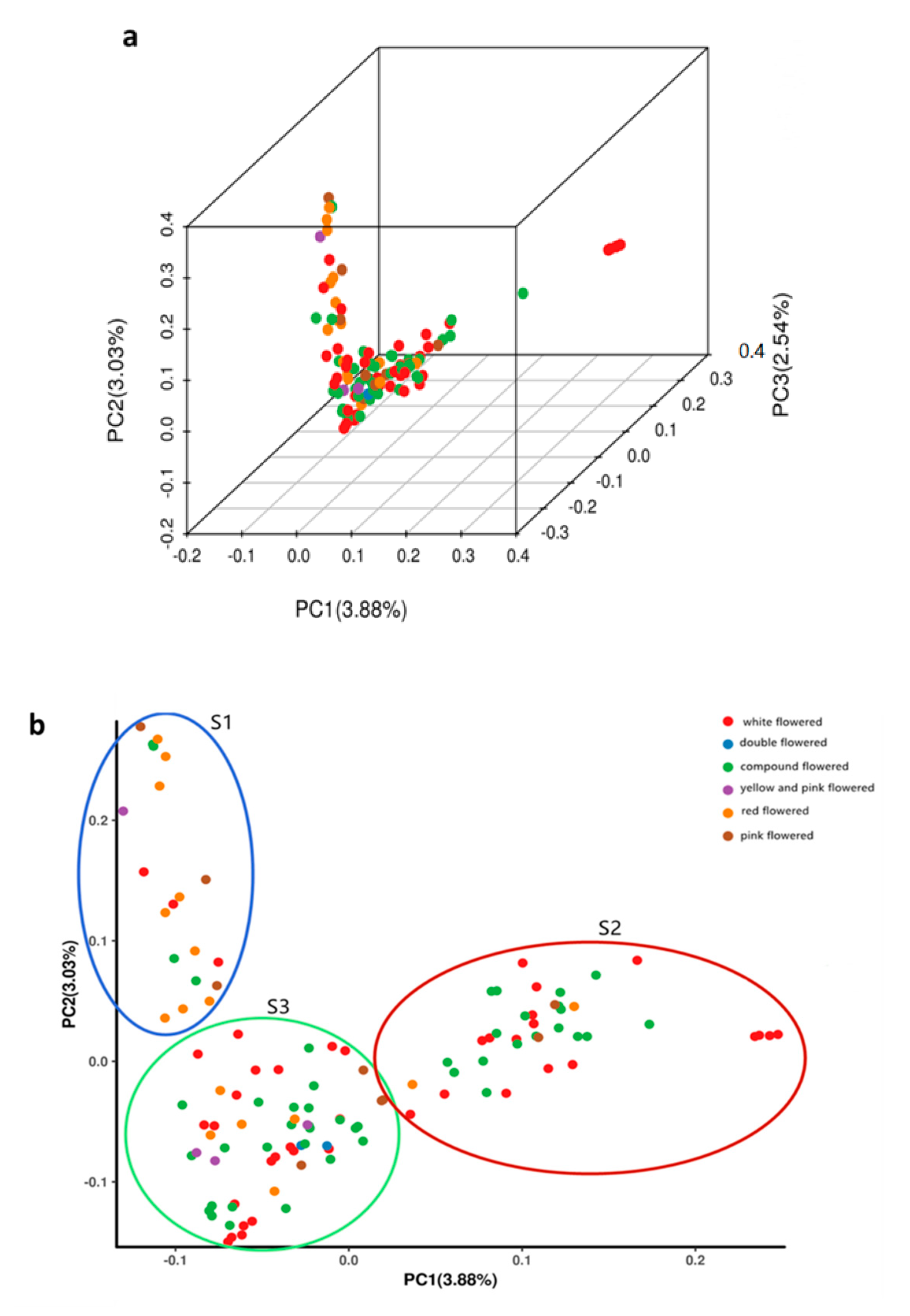
3.4. Principal Component Analysis
3.5. Genetic Diversity Analysis
3.6. Core Collection Establishment and Evaluation
4. Discussion
4.1. SNP-Based Genetic Relationships among the X. sorbifolium Germplasm Resources
4.2. Genetic Diversity of X. sorbifolium Germplasm Resources
4.3. Core Collection Establishment and Evaluation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mou, H.X.; Yu, H.Y.; Hou, X.C. Regular Distribution of Woody Energy Plant Xanthoceras sorbifolium Bunge in China. J. Anhui Agric. Sci. 2008, 36, 3626–3628. [Google Scholar]
- Shao, H.B.; Chu, L.Y. Resource evaluation of typical energy plants and possible functional zone planning in China. Biomass Bioenergy 2008, 32, 283–288. [Google Scholar] [CrossRef]
- Fu, Y.J.; Zu, Y.G.; Wang, L.L.; Zhang, N.J.; Liu, W.; Li, S.M.; Zhang, S. Determination of Fatty Acid Methyl Esters in Biodiesel Produced from Yellow Horn Oil by LC. Chromatographia 2008, 67, 9–14. [Google Scholar] [CrossRef]
- Zhao, Y.; Liu, X.J.; Wang, M.K.; Bi, Q.X.; Cui, Y.F.; Wang, L.B. Transcriptome and physiological analyses provide insights into the leaf epicuticular wax accumulation mechanism in yellowhorn. Hortic. Res. 2021, 8, 134–144. [Google Scholar] [CrossRef]
- Venegas-Calerón, M.; Ruíz-Méndez, M.V.; Martínez-Force, E. Characterization of Xanthoceras sorbifolium Bunge seeds: Lipids, proteins and saponins content. Ind. Crops Prod. 2017, 109, 192–198. [Google Scholar] [CrossRef] [Green Version]
- Bi, Q.X.; Zhao, Y.; Cui, Y.F.; Wang, L.B. Genome survey sequencing and genetic background characterization of yellow horn based on next-generation sequencing. Mol. Biol. Rep. 2019, 46, 4303–4312. [Google Scholar] [CrossRef]
- Rong, W.; Sun, Z.; Li, Q.; Liu, R.; Zhang, T.; Wang, T.; Yang, W.; Li, Z.; Bi, K. Characterization and simultaneous quantification of seven triterpenoid saponins in different parts of Xanthoceras sorbifolia Bunge by HPLC-ESI-TOF. Anal. Methods 2016, 8, 2176–2184. [Google Scholar] [CrossRef]
- Zhang, Y.; Xiao, L.; Xiao, B.; Yin, M.; Gu, M.Y.; Zhong, R.; Shang, Y.; Wang, K.; Wei, L. Research progress and application prospect of Xanthoceras sorbifolia for treating Alzheimer’s disease. Drug Eval. Res. 2018, 41, 912–917. [Google Scholar]
- Qi, Y.; Ji, X.F.; Chi, T.Y.; Liu, P.; Jin, G.; Xu, Q.; Jiao, Q.; Wang, L.H.; Zou, L.B. Xanthoceraside attenuates amyloid β peptide1-42-induced memory impairments by reducing neuroinflammatory responses in mice. Eur. J. Pharmacol. 2017, 820, 18–30. [Google Scholar] [CrossRef]
- Wang, Y.L.; Yue, S.L.; Li, R.F.; Zhai, H.X.; Song, Q. A New Cultivar of Landscaping Xanthoceras sorbifolium ‘Senmiao Jinziguan’. Acta Hortic. Sin. 2020, 47, 405–406. [Google Scholar]
- Zhou, Q.Y.; Cai, Q. The superoxide dismutase genes might be required for appropriate development of the ovule after fertilization in Xanthoceras sorbifolium. Plant Cell Rep. 2018, 37, 727–739. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.Y.; Fan, S.Q.; Bi, Q.X.; Wang, S.X.; Hu, X.Y.; Chen, M.Y.; Wang, L.B. Seed morphology, oil content and fatty acid composition variability assessment in yellow horn (Xanthoceras sorbifolium Bunge) germplasm for optimum biodiesel production. Ind. Crops Prod. 2017, 97, 425–430. [Google Scholar] [CrossRef]
- Ren, X.B.; Li, S.C.; Li, F.M.; Sun, J.H.; Zhang, L.M.; Li, X.G. The genetic germplasm collection field construction and sapling increment evaluation of Xanthoceras sorbifolia. J. Jilin For. Sci. Technol. 2016, 45, 21–25. [Google Scholar]
- Schafleitner, R.; Nair, R.M.; Rathore, A.; Wang, Y.W.; Lin, C.Y.; Chu, S.H.; Lin, P.Y.; Chang, J.C.; Ebert, A.W. The World Vegetable Center mungbean (Vigna radiata) core and mini core collections. BMC Genom. 2015, 16, 344. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.W.; Wu, S.Q.; Kou, Y.Y.; Luo, G.J. RAPD analysis of germplasm resources of Xanthoceras sorbifolia. J. Liaoning For. Sci. Technol. 2012, 1, 11–12. [Google Scholar]
- Guan, L.P.; Yang, T.; Li, N.; Li, B.S.; Lu, H. Identification of superior clones by RAPD technology in Xanthoceras sorbifolia Bunge. For. Stud. China 2010, 12, 37–40. [Google Scholar] [CrossRef]
- Bi, Q.X.; Guan, W.B. Isolation and characterisation of polymorphic genomic SSRs markers for the endangered tree Xanthoceras sorbifolium Bunge. Conserv. Genet. Resour. 2014, 6, 895–898. [Google Scholar] [CrossRef]
- Bi, Q.X.; Mao, J.F.; Guan, W.B. Efficiently developing a large set of polymorphic EST-SSR markers for Xanthoceras sorbifolium by mining raw reads from high-throughput sequencing. Conserv. Genet. Resour. 2015, 7, 423–425. [Google Scholar] [CrossRef]
- Shen, Z.; Zhang, K.; Ma, L.; Duan, J.; Ao, Y. Analysis of the genetic relationships and diversity among 11 populations of Xanthoceras sorbifolia using phenotypic and microsatellite marker data. Electron. J. Biotechnol. 2017, 26, 33–39. [Google Scholar] [CrossRef]
- Shen, Z.; Duan, J.; Ma, L.Y. Genetic diversity of Xanthoceras sorbifolium bunge germplasm using morphological traits and microsatellite molecular markers. PLoS ONE 2017, 7, e0177577. [Google Scholar] [CrossRef] [Green Version]
- Chanhoon, A.; Hyunseok, L.; Jeonghoon, L.; Eun, J.C.; Li, Y.H.; Jaeseon, Y. Analysis of genetic diversity and differentiation of artificial populations of yellowhorn (Xanthoceras sorbifolium) in China using ISSR markers. J. For. Res. 2016, 27, 1099–1104. [Google Scholar]
- El-Kassaby, Y.A.; Wang, Q.; Wang, T.; Ratcliffe, B.; Bi, Q.X.; Wang, Z.; Mao, J.F.; Guan, W.B. Concept for gene conservation strategy for the endangered Chinese yellowhorn, Xanthoceras sorbifolium, based on simulation of pairwise kinship coefficients. For. Ecol. Manag. 2019, 432, 976–982. [Google Scholar] [CrossRef]
- Wurschum, T.; Langer, S.M.; Longin, C.F.H. Population structure, genetic diversity and linkage disequilibrium in elite winter wheat assessed with SNP and SSR markers. Appl Genet 2013, 126, 1477–1486. [Google Scholar] [CrossRef]
- Verma, S.; Gupta, S.; Bandhiwal, N. High-density linkage map construction and mapping of seed trait QTLs in chickpea (Cicer arietinum L.) using genotyping-by-sequencing (GBS). Sci. Rep. 2015, 5, 17512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ai, X.; Liang, Y.; Wang, J.; Zheng, J.Z.; Gong, J.; Guo, X.; Qu, Y. Genetic Diversity and Structure of Elite Cotton Germplasm (Gossypium Hirsutum L.) Using Genome-Wide Snp Data. Genetica 2017, 145, 4–5. [Google Scholar] [CrossRef]
- Kumar, D.; Chhokar, V.; Sheoran, S.; Singh, R.; Sharma, P.; Jaiswal, S.; Iquebal, M.A. Characterization of Genetic Diversity and Population Structure in Wheat Using Array Based Snp Markers. Mol. Biol. Rep. 2020, 47, 293–306. [Google Scholar] [CrossRef]
- Yin, M.H.; Wang, Q.; Zhang, H.L.; Cai, X.H.; Xu, C.Q.; Chen, F.L.; Liu, S.Y.; Zhang, Q.W.; Ca, H.; Chen, R.H. Whole Genome Re-sequencing Analysis of Alpine Potato and Local Farm Potato in Huaiyu Mountain under High Altitude Habitats. Genom. Appl. Biol. 2020, 39, 1198–1207. [Google Scholar]
- Tan, Q.J.; Wang, W.L.; Chen, H.S.; Wei, Y.R.; Zheng, S.F.; Huang, X.Y.; He, P.; Xia, Z.Q. Genetic Diversity of Macadamia Varieties Based on Single-Nucleotide Polymorphism. Mol. Plant Breed. 2020, 18, 7246–7253. [Google Scholar]
- Bi, Q.X.; Zhao, Y.; Du, W.; Lu, Y.; Gui, L.L.; Zheng, Z.M.; Yu, H.Y.; Cui, Y.F.; Liu, Z.; Cui, T.P.; et al. Pseudomolecule-level assembly of the Chinese oil tree yellowhorn (Xanthoceras sorbifolium) genome. GigaScience 2019, 8, giz070. [Google Scholar] [CrossRef]
- Liang, Q.; Yang, H.; Li, S.k.; Yuan, F.L.; Sun, J.F.; Duan, Q.C.; Li, Q.Y.; Zhang, R.; Sang, Y.L.; Wang, N.; et al. The genome assembly and annotation of yellowhorn (Xanthoceras sorbifolium Bunge). GigaScience 2019, 8, giz071. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [Green Version]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 297–303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Picard. Available online: http://sourceforge.net/projects/picard/ (accessed on 29 December 2021).
- Yang, B.M.; Zhang, G.L.; Guo, F.P.; Wang, M.Q.; Wang, H.Y.; Xiao, H.X. A Genomewide Scan for Genetic Structure and Demographic History of Two Closely Related Species, Rhododendron dauricum and R. mucronulatum (Rhododendron, Ericaceae). Front. Plant Sci. 2020, 11, 1093. [Google Scholar] [CrossRef] [PubMed]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Liu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Alexander, D.H.; Novembre, J.; Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009, 19, 1655–1664. [Google Scholar] [CrossRef] [Green Version]
- Price, A.L.; Patterson, N.P. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006, 38, 904–909. [Google Scholar] [CrossRef]
- Thachuk, C.; Crossa, J.; Franco, J.; Dreisigacker, S.; Warburton, M.; Davenport, G.F. Core Hunter: An algorithm for sampling genetic resources based on multiple genetic measures. BMC Bioinform. 2009, 10, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Rafalski, A. Applications of single nucleotide polymorphisms in crop genetics. Curr. Opin. Plant Biol. 2002, 5, 94–100. [Google Scholar] [CrossRef]
- Mammadov, J.; Aggarwal, R.; Buyyarapu, R.; Kumpatla, S. SNP markers and their impact on plant breeding. Int. J. Plant Genom. 2012, 2012, 728398. [Google Scholar] [CrossRef]
- Jehan, T.; Lakhanpaul, S. Single nucleotide polymorphism (SNP)–methods and applications in plant genetics: A review. Indian J. Biotechnol. 2006, 5, 435–459. [Google Scholar]
- Wang, Y.Y.; Lv, H.K.; Xiang, X.H.; Yang, A.G.; Feng, Q.F.; Dai, P.G.; Li, Y.; Jiang, X.; Liu, G.X.; Zhang, X.W. Construction of a SNP Fingerprinting Database and Population Genetic Analysis of Cigar Tobacco Germplasm Resources in China. Front. Plant Sci. 2021, 12, 618133. [Google Scholar] [CrossRef] [PubMed]
- Qiao, D.Y.; Wang, P.; Wang, S.A.; Li, L.F.; Gao, L.L.; Yang, R.T.; Wang, Q.; Li, Y. Genetic Diversity Analysis of Lagerstroemia Germplasm Resources Based on SNP Markers. J. Nanjing For. Univ. 2020, 44, 21–28. [Google Scholar]
- Lang, Y.H.; Sun, Y.; Feng, Y.J.; Qi, Z.; Yu, M.; Song, K. Recent Progress in the Molecular Investigations of Yellow Horn (Xanthoceras sorbifolia Bunge). Bot. Rev. 2020, 86, 136–148. [Google Scholar] [CrossRef]
- Botstein, D.; White, R.L.; Skolnick, M.; Davis, R.W. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 1980, 32, 314–331. [Google Scholar]
- Eltaher, S.; Sallam, A.; Belamkar, V. Genetic diversity and population structure of F3:6 nebraska winter wheat geno-types using genotyping-by-sequencing. Front. Genet. 2018, 9, 76. [Google Scholar] [CrossRef]
- Alemu, A.; Feyissa, T.; Letta, T.; Abeyo, B. Genetic diversity and population structure analysis based on the high density SNP markers in Ethiopian durum wheat (Triticum turgidum ssp. durum). BMC Genet. 2020, 21, 18–35. [Google Scholar] [CrossRef] [Green Version]
- Nayak, S.N.; Song, J.; Villa, A.; Pathak, B.; Ayala, S.T.; Yang, X.P.; Todd, J.; Glynn, N.C.; Kuhn, D.N.; Glaz, B.; et al. Promoting Utilization of Saccharum spp. Genetic Resources through Genetic Diversity Analysis and Core Collection Construction. PLoS ONE 2014, 9, e110856. [Google Scholar] [CrossRef] [Green Version]
- Yonezawa, K.; Nomura, T.; Morishima, H. Sampling strategies for use in stratified germplasm collections. In Core Collections of Plant Genetic Resources; Hodgkin, T., Brown, A.H.D., Van Hintum, T.J.L., Eds.; John Wiley & Sons: Chichester, UK, 1995; pp. 35–53. [Google Scholar]
- Agrama, H.A.; Yan, W.G.; Lee, F.; Fjellstrom, R.; Chen, M.H. Genetic assessment of a mini-core subset developed from USDA Rice Genebank. Crop Sci. 2009, 49, 1336–1346. [Google Scholar] [CrossRef]
- Shen, Z.; Ma, L.Y.; Ao, Y.; Duan, J. Analysis of the Genetic Diversity and Construction of Core Collection of Xanthoceras sorbifolia Bunge. Using Microsatellite Marker Data. Mol. Plant Breed. 2017, 15, 3341–3350. [Google Scholar]
- Xu, C.Q.; Gao, J.; Du, Z.F.; Li, D.K.; Wang, Z.; Li, Y.Y.; Pang, X.M. Identifying the genetic diversity, genetic structure and a core collection of Ziziphus jujube Mill. var. jujuba accessions using microsatellite markers. Sci. Rep. 2016, 6, 31503. [Google Scholar] [PubMed]
| Group | No. | MAF | Ne | He | H | A | Na | Ho | PIC | I |
|---|---|---|---|---|---|---|---|---|---|---|
| Q1 | 19 | 0.2157 ± 0.1267 | 1.4948 ± 0.2876 | 0.3063 ± 0.1303 | 0.3155 ± 0.1343 | 535,470 | 2 | 0.2115 ± 0.1446 | 0.2509 ± 0.0912 | 0.4734 ± 0.1569 |
| Q2 | 38 | 0.2045 ± 0.1194 | 1.4687 ± 0.2719 | 0.2968 ± 0.1218 | 0.3014 ± 0.1237 | 677,676 | 2 | 0.1539 ± 0.1033 | 0.2454 ± 0.0846 | 0.4638 ± 0.1454 |
| Q3 | 62 | 0.1950 ± 0.1186 | 1.4468 ± 0.2719 | 0.2858 ± 0.1237 | 0.2886 ± 0.1248 | 788,843 | 2 | 0.1649 ± 0.1016 | 0.2373 ± 0.0865 | 0.4501 ± 0.1486 |
| Average/Total | 119 | 0.2051 ± 0.1190 | 1.4701 ± 0.2737 | 0.2963 ± 0.1276 | 0.3018 ± 0.1282 | 667,330 | 2 | 0.1768 ± 0.0896 | 0.2445 ± 0.0905 | 0.4624 ± 0.1552 |
| White-flowered | 42 | 0.1904 ± 0.1174 | 1.4360 ± 0.2694 | 0.2808 ± 0.1234 | 0.2847 ± 0.1251 | 751,227 | 2 | 0.1528 ± 0.1039 | 0.2337 ± 0.0867 | 0.4439 ± 0.1487 |
| Double flowered | 2 | 0.3500 ± 0.1225 | 1.7600 ± 0.1960 | 0.4250 ± 0.0612 | 0.5667 ± 0.0816 | 282,886 | 2 | 0.3766 ± 0.2911 | 0.3328 ± 0.0344 | 0.6147 ± 0.0641 |
| Compound-flowered | 46 | 0.1896 ± 0.1184 | 1.4335 ± 0.2713 | 0.2792 ± 0.1241 | 0.2828 ± 0.1257 | 874,440 | 2 | 0.1523 ± 0.1003 | 0.2325 ± 0.0871 | 0.4419 ± 0.1495 |
| Yellow- and pink-flowered | 4 | 0.2581 ± 0.1247 | 1.5874 ± 0.2664 | 0.3519 ± 0.1088 | 0.4022 ± 0.1244 | 333,550 | 2 | 0.2722 ± 0.2074 | 0.2841 ± 0.0714 | 0.5304 ± 0.1240 |
| Red-flowered | 16 | 0.2103 ± 0.1234 | 1.4817 ± 0.2799 | 0.3017 ± 0.1250 | 0.3128 ± 0.1296 | 657,812 | 2 | 0.1755 ± 0.1341 | 0.2484 ± 0.0866 | 0.4692 ± 0.1489 |
| Pink-flowered | 9 | 0.2092 ± 0.1274 | 1.4789 ± 0.2882 | 0.2985 ± 0.1317 | 0.3174 ± 0.1401 | 560,379 | 2 | 0.1871 ± 0.1491 | 0.2453 ± 0.0927 | 0.4638 ± 0.1594 |
| Average/Total | 119 | 0.2346 ± 0.1190 | 1.5296 ± 0.2737 | 0.3229 ± 0.1276 | 0.3611 ± 0.1282 | 576,715 | 2 | 0.2194 ± 0.0896 | 0.2628 ± 0.0905 | 0.4940 ± 0.1552 |
| States | Number of Germplasms | Ho | He | H | I | PIC |
|---|---|---|---|---|---|---|
| All | 119 | 0.008–1.000 (0.142) | 0.095–0.500 (0.266) | 0.095–0.503 (0.268) | 0.199–0.693 (0.425) | 0.090–0.375 (0.223) |
| Core | 35 | 0.029–1.000 (0.152) | 0.095–0.500 (0.288) | 0.097–0.509 (0.293) | 0.199–0.693 (0.453) | 0.090–0.375 (0.239) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Li, Y. Population Genetics and Development of a Core Collection from Elite Germplasms of Xanthoceras sorbifolium Based on Genome-Wide SNPs. Forests 2022, 13, 338. https://doi.org/10.3390/f13020338
Wang Y, Li Y. Population Genetics and Development of a Core Collection from Elite Germplasms of Xanthoceras sorbifolium Based on Genome-Wide SNPs. Forests. 2022; 13(2):338. https://doi.org/10.3390/f13020338
Chicago/Turabian StyleWang, Yali, and Yi Li. 2022. "Population Genetics and Development of a Core Collection from Elite Germplasms of Xanthoceras sorbifolium Based on Genome-Wide SNPs" Forests 13, no. 2: 338. https://doi.org/10.3390/f13020338





