Construction of Core Collection and Phenotypic Evaluation of Toona sinensis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Experimental Methods
2.2.1. DNA Extraction
2.2.2. SSR Typing
2.2.3. Measurement of Phenotypic Traits
2.3. Data Analysis
2.3.1. Analysis of the Genetic Diversity and Structure of the Breeding Population
2.3.2. Construction of Core Collection
2.3.3. Analysis of Phenotypic Data
3. Results
3.1. Analysis of Genetic Diversity of Breeding Population
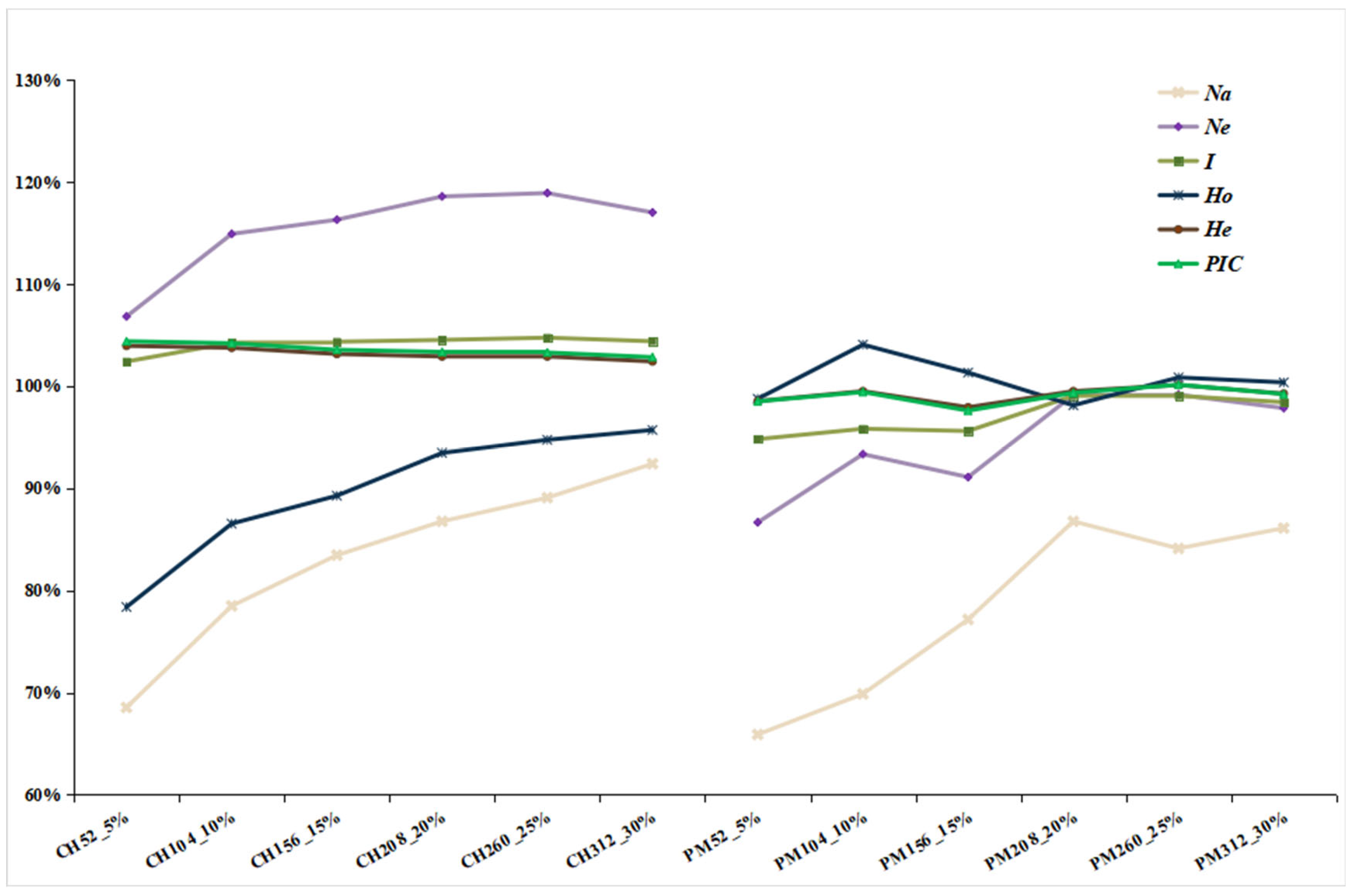
3.2. Construction of Alternative Core Collection
3.3. Genetic Structure Analysis of the T. sinensis Breeding Population
3.4. Comprehensive Evaluation and Analysis of Core Collection
3.4.1. Descriptive Statistics
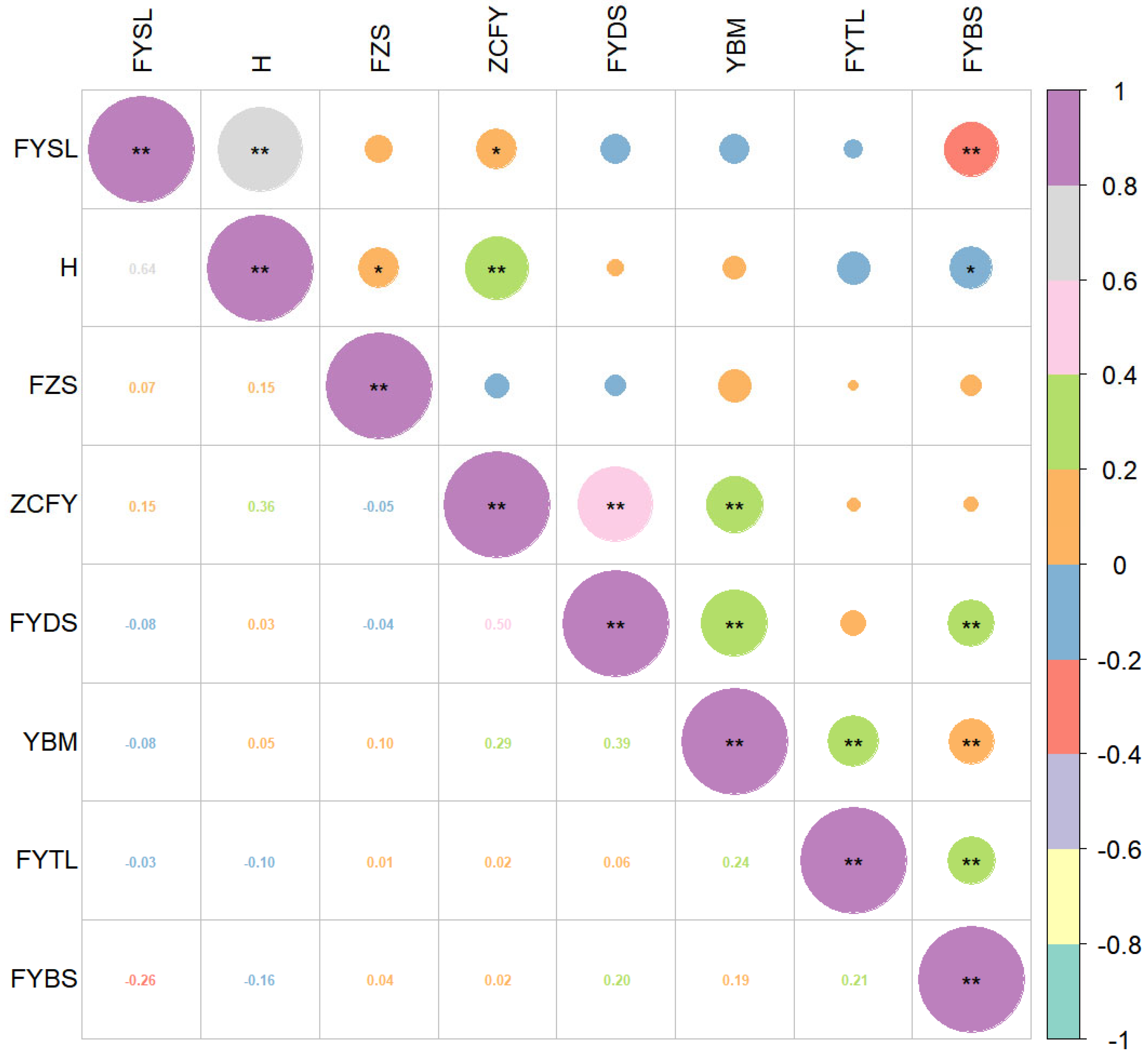
3.4.2. Correlation Analysis of Eight Agronomic Traits
3.4.3. Weight Determination of Each Trait Based on PCA
3.4.4. Comprehensive Evaluation of Core Collection Based on the TOPSIS Method
4. Discussion
4.1. Construction of Core Collection
4.2. Comprehensive Evaluation of Multiple Traits of Individual Plants in the Core Collection
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Appendix A
| Sampling Method | Na | Ne | I | Ho | He | PIC |
| CH52_5% | 15.923 | 8.668 | 2.317 | 0.487 | 0.856 | 0.841 |
| CH104_10% | 18.231 | 9.324 | 2.359 | 0.538 | 0.854 | 0.839 |
| CH156_15% | 19.385 | 9.437 | 2.361 | 0.555 | 0.849 | 0.834 |
| CH208_20% | 20.154 | 9.622 | 2.365 | 0.581 | 0.847 | 0.833 |
| CH260_25% | 20.692 | 9.649 | 2.370 | 0.589 | 0.847 | 0.832 |
| CH312_30% | 21.462 | 9.495 | 2.362 | 0.595 | 0.843 | 0.828 |
| PM52_5% | 15.308 | 7.031 | 2.145 | 0.614 | 0.811 | 0.794 |
| PM104_10% | 16.231 | 7.573 | 2.168 | 0.647 | 0.819 | 0.801 |
| PM156_15% | 17.923 | 7.391 | 2.163 | 0.630 | 0.806 | 0.786 |
| PM208_20% | 20.154 | 8.043 | 2.241 | 0.610 | 0.819 | 0.800 |
| PM260_25% | 19.538 | 8.046 | 2.241 | 0.627 | 0.824 | 0.807 |
| PM312_30% | 20.000 | 7.939 | 2.227 | 0.624 | 0.817 | 0.799 |
| 1040_100% | 23.231 | 8.114 | 2.263 | 0.622 | 0.823 | 0.806 |
Appendix B
| Number | Rank | Relative Proximity of the Ideal Solution (Ci) | >Positive Ideal Solutions (X+) | Negative Ideal Solutions (X−) |
| N0237 | 1 | 0.734 | 0.241 | 0.667 |
| N0972 | 2 | 0.725 | 0.252 | 0.663 |
| N0409 | 3 | 0.711 | 0.261 | 0.642 |
| N0783 | 4 | 0.707 | 0.279 | 0.673 |
| N0048 | 5 | 0.696 | 0.28 | 0.641 |
| N0802 | 6 | 0.683 | 0.3 | 0.646 |
| N0845 | 7 | 0.676 | 0.31 | 0.647 |
| N0981 | 8 | 0.675 | 0.317 | 0.659 |
| N0125 | 9 | 0.665 | 0.323 | 0.641 |
| N0074 | 10 | 0.664 | 0.325 | 0.642 |
| N0989 | 11 | 0.663 | 0.321 | 0.631 |
| N0064 | 12 | 0.663 | 0.35 | 0.688 |
| N0300 | 13 | 0.661 | 0.314 | 0.612 |
| N0165 | 14 | 0.66 | 0.317 | 0.616 |
| N0556 | 15 | 0.659 | 0.346 | 0.669 |
| N0397 | 16 | 0.657 | 0.334 | 0.642 |
| N1002 | 17 | 0.656 | 0.323 | 0.616 |
| N0569 | 18 | 0.656 | 0.335 | 0.638 |
| N0814 | 19 | 0.656 | 0.316 | 0.601 |
| N0750 | 20 | 0.653 | 0.328 | 0.619 |
| N0820 | 21 | 0.65 | 0.331 | 0.616 |
| N0842 | 22 | 0.646 | 0.336 | 0.612 |
| N0002 | 23 | 0.644 | 0.337 | 0.61 |
| N0986 | 24 | 0.641 | 0.323 | 0.577 |
| N0192 | 25 | 0.641 | 0.352 | 0.63 |
| N0844 | 26 | 0.641 | 0.351 | 0.626 |
| N0082 | 27 | 0.64 | 0.351 | 0.622 |
| N0424 | 28 | 0.638 | 0.329 | 0.581 |
| N0522 | 29 | 0.638 | 0.332 | 0.585 |
| N0068 | 30 | 0.636 | 0.342 | 0.599 |
| N0303 | 31 | 0.633 | 0.351 | 0.604 |
| N0525 | 32 | 0.632 | 0.345 | 0.591 |
| N0853 | 33 | 0.631 | 0.371 | 0.634 |
| N0326 | 34 | 0.63 | 0.342 | 0.583 |
| N0123 | 35 | 0.628 | 0.361 | 0.611 |
| N0324 | 36 | 0.625 | 0.36 | 0.6 |
| N0041 | 37 | 0.625 | 0.361 | 0.602 |
| N0053 | 38 | 0.624 | 0.368 | 0.611 |
| N0987 | 39 | 0.624 | 0.399 | 0.662 |
| N0831 | 40 | 0.624 | 0.365 | 0.605 |
| N0792 | 41 | 0.622 | 0.371 | 0.61 |
| N0065 | 42 | 0.621 | 0.371 | 0.608 |
| N0701 | 43 | 0.62 | 0.385 | 0.628 |
| N0352 | 44 | 0.616 | 0.333 | 0.536 |
| N0650 | 45 | 0.616 | 0.385 | 0.618 |
| N0157 | 46 | 0.615 | 0.394 | 0.63 |
| N0927 | 47 | 0.613 | 0.381 | 0.604 |
| N0866 | 48 | 0.613 | 0.362 | 0.573 |
| N0904 | 49 | 0.61 | 0.385 | 0.603 |
| N0992 | 50 | 0.61 | 0.357 | 0.559 |
| N0455 | 51 | 0.609 | 0.366 | 0.57 |
| N0765 | 52 | 0.608 | 0.376 | 0.583 |
| N0418 | 53 | 0.606 | 0.39 | 0.599 |
| N0240 | 54 | 0.606 | 0.363 | 0.558 |
| N0143 | 55 | 0.604 | 0.379 | 0.578 |
| N0136 | 56 | 0.604 | 0.392 | 0.597 |
| N0983 | 57 | 0.604 | 0.397 | 0.605 |
| N0079 | 58 | 0.603 | 0.378 | 0.575 |
| N0795 | 59 | 0.603 | 0.384 | 0.584 |
| N0381 | 60 | 0.603 | 0.407 | 0.619 |
| N0826 | 61 | 0.603 | 0.376 | 0.571 |
| N0139 | 62 | 0.602 | 0.372 | 0.563 |
| N0859 | 63 | 0.6 | 0.39 | 0.585 |
| N0641 | 64 | 0.599 | 0.382 | 0.571 |
| N0512 | 65 | 0.599 | 0.405 | 0.604 |
| N0439 | 66 | 0.598 | 0.381 | 0.568 |
| N0539 | 67 | 0.598 | 0.395 | 0.587 |
| N0340 | 68 | 0.597 | 0.396 | 0.587 |
| N0977 | 69 | 0.595 | 0.382 | 0.561 |
| N0863 | 70 | 0.594 | 0.409 | 0.6 |
| N0290 | 71 | 0.592 | 0.411 | 0.596 |
| N0793 | 72 | 0.592 | 0.375 | 0.543 |
| N0364 | 73 | 0.59 | 0.371 | 0.533 |
| N0189 | 74 | 0.589 | 0.403 | 0.577 |
| N0248 | 75 | 0.588 | 0.399 | 0.569 |
| N0103 | 76 | 0.588 | 0.375 | 0.534 |
| N0769 | 77 | 0.587 | 0.396 | 0.564 |
| N0150 | 78 | 0.587 | 0.378 | 0.537 |
| N0329 | 79 | 0.585 | 0.396 | 0.558 |
| N0865 | 80 | 0.583 | 0.389 | 0.544 |
| N0764 | 81 | 0.582 | 0.403 | 0.56 |
| N0119 | 82 | 0.581 | 0.421 | 0.585 |
| N0159 | 83 | 0.581 | 0.425 | 0.59 |
| N0587 | 84 | 0.58 | 0.405 | 0.561 |
| N0834 | 85 | 0.578 | 0.417 | 0.572 |
| N0114 | 86 | 0.577 | 0.403 | 0.549 |
| N0991 | 87 | 0.575 | 0.405 | 0.548 |
| N0272 | 88 | 0.573 | 0.404 | 0.542 |
| N0663 | 89 | 0.572 | 0.41 | 0.548 |
| N0912 | 90 | 0.571 | 0.432 | 0.576 |
| N0564 | 91 | 0.571 | 0.42 | 0.559 |
| N0388 | 92 | 0.57 | 0.412 | 0.546 |
| N0784 | 93 | 0.57 | 0.442 | 0.587 |
| N0604 | 94 | 0.565 | 0.429 | 0.558 |
| N0878 | 95 | 0.565 | 0.416 | 0.54 |
| N0787 | 96 | 0.564 | 0.426 | 0.551 |
| N0608 | 97 | 0.564 | 0.443 | 0.574 |
| N0152 | 98 | 0.564 | 0.451 | 0.584 |
| N0858 | 99 | 0.562 | 0.427 | 0.547 |
| N0317 | 100 | 0.562 | 0.433 | 0.555 |
| N0872 | 101 | 0.562 | 0.452 | 0.579 |
| N0244 | 102 | 0.561 | 0.419 | 0.535 |
| N0190 | 103 | 0.56 | 0.427 | 0.543 |
| N0861 | 104 | 0.559 | 0.411 | 0.521 |
| N0821 | 105 | 0.557 | 0.396 | 0.498 |
| N0530 | 106 | 0.556 | 0.448 | 0.56 |
| N1013 | 107 | 0.555 | 0.424 | 0.529 |
| N0613 | 108 | 0.555 | 0.419 | 0.522 |
| N0854 | 109 | 0.554 | 0.432 | 0.537 |
| N0030 | 110 | 0.554 | 0.437 | 0.543 |
| N0115 | 111 | 0.553 | 0.421 | 0.521 |
| N0689 | 112 | 0.552 | 0.434 | 0.534 |
| N0277 | 113 | 0.551 | 0.437 | 0.536 |
| N0603 | 114 | 0.55 | 0.429 | 0.524 |
| N0444 | 115 | 0.549 | 0.433 | 0.528 |
| N0448 | 116 | 0.548 | 0.406 | 0.491 |
| N0760 | 117 | 0.547 | 0.429 | 0.519 |
| N0018 | 118 | 0.541 | 0.478 | 0.564 |
| N1028 | 119 | 0.541 | 0.507 | 0.597 |
| N0615 | 120 | 0.54 | 0.447 | 0.525 |
| N0763 | 121 | 0.539 | 0.455 | 0.533 |
| N0505 | 122 | 0.538 | 0.499 | 0.582 |
| N0940 | 123 | 0.538 | 0.468 | 0.546 |
| N0901 | 124 | 0.536 | 0.46 | 0.532 |
| N1001 | 125 | 0.535 | 0.468 | 0.539 |
| N0009 | 126 | 0.534 | 0.456 | 0.523 |
| N0685 | 127 | 0.532 | 0.438 | 0.498 |
| N0906 | 128 | 0.531 | 0.488 | 0.553 |
| N0852 | 129 | 0.531 | 0.425 | 0.48 |
| N0026 | 130 | 0.53 | 0.459 | 0.517 |
| N0789 | 131 | 0.528 | 0.447 | 0.501 |
| N0681 | 132 | 0.528 | 0.443 | 0.495 |
| N0016 | 133 | 0.527 | 0.506 | 0.565 |
| N0748 | 134 | 0.527 | 0.433 | 0.482 |
| N0761 | 135 | 0.526 | 0.451 | 0.501 |
| N0242 | 136 | 0.526 | 0.476 | 0.527 |
| N0653 | 137 | 0.525 | 0.442 | 0.488 |
| N0855 | 138 | 0.522 | 0.456 | 0.499 |
| N0662 | 139 | 0.522 | 0.42 | 0.458 |
| N0047 | 140 | 0.521 | 0.456 | 0.496 |
| N0917 | 141 | 0.521 | 0.462 | 0.503 |
| N0598 | 142 | 0.519 | 0.455 | 0.491 |
| N0147 | 143 | 0.518 | 0.456 | 0.489 |
| N0728 | 144 | 0.517 | 0.535 | 0.573 |
| N0741 | 145 | 0.517 | 0.469 | 0.502 |
| N0879 | 146 | 0.515 | 0.458 | 0.487 |
| N0096 | 147 | 0.514 | 0.494 | 0.523 |
| N0172 | 148 | 0.514 | 0.531 | 0.56 |
| N0033 | 149 | 0.513 | 0.473 | 0.498 |
| N0450 | 150 | 0.512 | 0.43 | 0.452 |
| N0737 | 151 | 0.511 | 0.482 | 0.505 |
| N0085 | 152 | 0.51 | 0.447 | 0.465 |
| N0284 | 153 | 0.508 | 0.475 | 0.49 |
| N0066 | 154 | 0.505 | 0.47 | 0.479 |
| N0058 | 155 | 0.504 | 0.5 | 0.507 |
| N0116 | 156 | 0.503 | 0.452 | 0.457 |
| N0993 | 157 | 0.5 | 0.465 | 0.466 |
| N0267 | 158 | 0.499 | 0.458 | 0.456 |
| N0828 | 159 | 0.498 | 0.467 | 0.463 |
| N0377 | 160 | 0.497 | 0.485 | 0.48 |
| N0875 | 161 | 0.49 | 0.479 | 0.461 |
| N0611 | 162 | 0.49 | 0.464 | 0.446 |
| N0563 | 163 | 0.489 | 0.522 | 0.5 |
| N0057 | 164 | 0.487 | 0.467 | 0.444 |
| N0752 | 165 | 0.486 | 0.506 | 0.479 |
| N0683 | 166 | 0.484 | 0.479 | 0.449 |
| N0708 | 167 | 0.483 | 0.504 | 0.472 |
| N0677 | 168 | 0.483 | 0.492 | 0.46 |
| N0269 | 169 | 0.482 | 0.47 | 0.438 |
| N0289 | 170 | 0.479 | 0.532 | 0.489 |
| N0963 | 171 | 0.479 | 0.578 | 0.531 |
| N0211 | 172 | 0.478 | 0.467 | 0.429 |
| N0909 | 173 | 0.475 | 0.555 | 0.502 |
| N0433 | 174 | 0.475 | 0.524 | 0.473 |
| N0900 | 175 | 0.471 | 0.563 | 0.501 |
| N0876 | 176 | 0.469 | 0.502 | 0.443 |
| N0740 | 177 | 0.469 | 0.522 | 0.461 |
| N0210 | 178 | 0.464 | 0.52 | 0.45 |
| N0944 | 179 | 0.464 | 0.593 | 0.513 |
| N0908 | 180 | 0.464 | 0.487 | 0.421 |
| N0145 | 181 | 0.462 | 0.479 | 0.411 |
| N0922 | 182 | 0.461 | 0.551 | 0.472 |
| N0915 | 183 | 0.461 | 0.527 | 0.451 |
| N0935 | 184 | 0.461 | 0.527 | 0.45 |
| N1021 | 185 | 0.459 | 0.494 | 0.419 |
| N0883 | 186 | 0.451 | 0.557 | 0.458 |
| N0371 | 187 | 0.449 | 0.505 | 0.411 |
| N0268 | 188 | 0.447 | 0.517 | 0.418 |
| N0137 | 189 | 0.445 | 0.508 | 0.406 |
| N0445 | 190 | 0.444 | 0.499 | 0.399 |
| N0091 | 191 | 0.442 | 0.523 | 0.414 |
| N0707 | 192 | 0.439 | 0.508 | 0.397 |
| N0434 | 193 | 0.439 | 0.492 | 0.384 |
| N0958 | 194 | 0.438 | 0.621 | 0.485 |
| N0744 | 195 | 0.436 | 0.609 | 0.471 |
| N0118 | 196 | 0.434 | 0.513 | 0.394 |
| N0664 | 197 | 0.432 | 0.58 | 0.441 |
| N0494 | 198 | 0.431 | 0.532 | 0.403 |
| N0032 | 199 | 0.426 | 0.541 | 0.402 |
| N0247 | 200 | 0.421 | 0.546 | 0.398 |
| N0903 | 201 | 0.419 | 0.558 | 0.402 |
| N0937 | 202 | 0.413 | 0.574 | 0.403 |
| N0962 | 203 | 0.405 | 0.567 | 0.386 |
| N0824 | 204 | 0.402 | 0.571 | 0.384 |
| N0218 | 205 | 0.4 | 0.567 | 0.378 |
| N0051 | 206 | 0.4 | 0.57 | 0.38 |
| N0036 | 207 | 0.384 | 0.589 | 0.367 |
| N0196 | 208 | 0.379 | 0.58 | 0.355 |
References
- Edmonds, J.M.; Staniforth, M. Toona sinensis: Meliaceae. Curtis’s Bot. Mag. 1998, 15, 186–193. [Google Scholar] [CrossRef]
- Hu, J.W.; Ma, W.J.; Shen, Y.Q.; Xiao, Y. Study on Growth Traits of Different Toona sinensis Clones and Superior Clone Early Selection. For. Res. 2019, 32, 165–170. [Google Scholar]
- Wei, P.; Yujie, L.; Meibian, H.; Mengmeng, Z.; Jing, Y.; Fang, L.; Qinwan, H.; Chunjie, W. Toona sinensis: A comprehensive review on its traditional usages, phytochemisty, pharmacology and toxicology. Rev. Bras. De Farmacogn. 2019, 29, 111–124. [Google Scholar]
- Hu, J.; Song, Y.; Mao, X.; Wang, Z.; Zhao, Q. Limonoids isolated from Toona sinensis and their radical scavenging, anti-inflammatory and cytotoxic activities. J. Funct. Foods 2016, 20, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Juan-Juan, C.; Qing-Qing, L.; Bao, Z.; Han-Qing, C. Structural characterization and hepatoprotective activities of polysaccharides from the leaves of Toona sinensis (A. Juss) Roem. Carbohydr. Polym. 2019, 212, 89–101. [Google Scholar]
- Chen, H.; Wu, Y.; Chia, Y.; Chang, F.; Hsu, H.; Hsieh, Y.; Chen, C.; Yuan, S. Gallic acid, a major component of Toona sinensis leaf extracts, contains a ROS-mediated anti-cancer activity in human prostate cancer cells. Cancer Lett. 2009, 286, 161–171. [Google Scholar] [CrossRef]
- Chen, H.; Wu, Y.; Chia, Y.; Chang, F.; Hsu, H.; Hsieh, Y.; Chen, C.; Yuan, S. Toona sinensis Roem tender leaf extract inhibits SARS coronavirus replication. J. Ethnopharmacol. 2008, 120, 108–111. [Google Scholar] [CrossRef]
- Li, Y.C.; Fan, Z.P.; Yang, J.; Liu, T.F. Research progress on functional components and biological activities of wild edible vegetables. J. Zhejiang AF Univ. 2022, 39, 913–922. [Google Scholar]
- Zhou, C.Y.; Ruan, J.Y.; Huang, J.; Tang, J. Advances in Research on Chemical Constituents and Biological Activities of Toona sinensis. Chin. Tradit. Pat. Med. 2020, 42, 1279–1291. [Google Scholar]
- Li, C.C. Study on Flavonoid Content and Antioxidant Properties of Toona sinensis Old Leaves. Food Res. Dev. 2021, 42, 34–39. [Google Scholar]
- Liu, Y.M.; Zhang, J.J.; Wu, L. Extraction of Total Flavonoids from Toona sinensis Old Leaves by Complex Enzymolysis Assisted with Ultrasound. Mod. Food Sci. Technol. 2019, 35, 223–230. [Google Scholar]
- Chen, W.; Li, C.C.; Ran, H.; Li, W.H. Isolation and Identification of Flavonoids and Saponins in Old Toona sinensis Leaves. Packag. Eng. 2019, 40, 36–42. [Google Scholar]
- Geng, Y.H.; Xu, X.; Ni, J.; Su, S. Effect of Harvest Time on Forage Quality of Toona sinensis. For. Res. 2019, 32, 145–151. [Google Scholar]
- Flora of China. Toona sinensis [Online Application Software]. Available online: http://www.efloras.org (accessed on 17 April 2023).
- Chen, Q.Q. Genetic Diversity Analysis of Toona sinensis Germplasm Resources in China; Shandong Agricultural University: Taian, China, 2018. [Google Scholar]
- Xu, X.; Guo, C.; Ma, C.; Li, M.; Chen, Y.; Liu, C.; Chu, J.; Yao, X. Brassinolide Soaking Reduced Nitrite Content and Extended Color Change and Storage Time of Toona sinensis Bud during Low Temperature and Near Freezing-Point Temperature Storage. Int. J. Mol. Sci. 2022, 23, 1311–1327. [Google Scholar] [CrossRef]
- Hseu, Y.; Chen, S.; Lin, W.; Hung, D.; Lin, M.; Kuo, Y.; Wang, M.; Cho, H.; Wang, L.; Yang, H. Toona sinensis (leaf extracts) inhibit vascular endothelial growth factor (VEGF)-induced angiogenesis in vascular endothelial cells. J. Ethnopharmacol. 2011, 134, 111–121. [Google Scholar] [CrossRef]
- Wang, X.; Xiao, Y.; He, Z.; Li, L.; Lv, Y.; Hu, X. Evolutionary Divergence between Toona ciliata and Toona sinensis Assayed with Their Whole Genome Sequences. Genes 2022, 13, 1799–1813. [Google Scholar] [CrossRef]
- Ji, Y.; Xiu, Z.; Chen, C.; Wang, Y.; Yang, J.; Sui, J.; Jiang, S.; Wang, P.; Yue, S.; Zhang, Q.; et al. Long read sequencing of Toona sinensis (A. Juss) Roem: A chromosome-level reference genome for the family Meliaceae. Mol. Ecol. Resour. 2021, 21, 1243–1255. [Google Scholar] [CrossRef]
- Liu, W.; Li, Y.; Liu, J.; Jiang, J. Estimation of Plant Height and Aboveground Biomass of Toona sinensis under Drought Stress Using RGB-D Imaging. Forests 2021, 12, 1747–1759. [Google Scholar] [CrossRef]
- Liu, W.; Li, Y.; Tomasetto, F.; Yan, W.; Tan, Z.; Liu, J.; Jiang, J. Non-destructive Measurements of Toona sinensis Chlorophyll and Nitrogen Content Under Drought Stress Using Near Infrared Spectroscopy. Front. Plant Sci. 2022, 12, 3274–3286. [Google Scholar] [CrossRef]
- Yao, C.C.; Wang, J.C.; Hu, J.W.; Xiao, Y. Genetic variation of growth and leaf phenotypic traits of Toona sinensis (A. Juss.) Roem. germplasms. Plant Sci. J. 2020, 38, 112–122. [Google Scholar]
- Wu, J.; Zhong, Z.Z.; Lou, J.; Yu, F. A Study on Phenotypic Diversity of Cone and Seed in Natural Populations of Toona sinensis. Acta Agric. Univ. Jiangxiensis 2018, 40, 248–256. [Google Scholar]
- Fu, D.; Zhong, K.; Zhong, Z.; Hu, G.; Zhang, P.; Tong, H. Genome-Wide Association Study of Sheath Blight Resistance within a Core Collection of Rice (Oryza sativa L.). Agronomy 2022, 12, 1493–1506. [Google Scholar] [CrossRef]
- Shi, A.; Gepts, P.; Song, Q.; Xiong, H.; Michaels, T.E.; Chen, S. Genome-Wide Association Study and Genomic Prediction for Soybean Cyst Nematode Resistance in USDA Common Bean (Phaseolus vulgaris) Core Collection. Front. Plant Sci. 2021, 12, 624–650. [Google Scholar] [CrossRef]
- Li, X.; Cui, L.; Zhang, L.; Huang, Y.; Zhang, S.; Chen, W.; Deng, X.; Jiao, Z.; Yang, W.; Qiu, Z. Genetic Diversity Analysis and Core Germplasm Collection Construction of Radish Cultivars Based on Structure Variation Markers. Int. J. Mol. Sci. 2023, 24, 2554–2566. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Liu, J.; Li, J.; Cao, S.; Zhang, Z.; Zhang, J.; Zhang, Y.; Deng, Y.; Niu, D.; Su, L.; et al. Genetic diversity and core collection extraction of Robinia pseudoacacia L. germplasm resources based on phenotype, physiology, and genotyping markers. Ind. Crops Prod. 2022, 178, 114–128. [Google Scholar] [CrossRef]
- Ortiz, R.; Ruiz TE, N.; Mujica, S.A. Sampling strategy for a core collection of Peruvian quinoa germplasm. TAG. Theoretical and applied genetics. Theor. Und Angew. Genet. 1998, 96, 75–83. [Google Scholar]
- Marcelo, F.O.; Randall, L.N.; Isaias, O.G.; Cosme, D.C.; José, F.F.D.T. Establishing a soybean germplasm core collection. Field Crops Res. 2010, 119, 277–289. [Google Scholar]
- Cretazzo, E.; Moreno, S.P.; Lorenzi, S.; Benítez, M.L.; Velasco, L.; Emanuelli, F. Genetic Characterization by SSR Markers of a Comprehensive Wine Grape Collection Conserved at Rancho de la Merced (Andalusia, Spain). Plants 2022, 11, 1088–1107. [Google Scholar] [CrossRef]
- Zhu, Y.Z.; Liang, D.Y.; Song, Z.J.; Tan, Y.; Guo, X.L.; Wang, D.L. Genetic Diversity Analysis and Core Germplasm Collection Construction of Camellia oleifera Based on Fruit Phenotype and SSR Data. Genes 2022, 13, 2351–2365. [Google Scholar] [CrossRef]
- Lv, J.; Li, C.; Zhou, C.; Chen, J.; Li, F.; Weng, Q.; Li, M.; Wang, Y.; Chen, S.; Chen, J. Genetic diversity analysis of a breeding population of Eucalyptus cloeziana F. Muell.(Myrtaceae) and extraction of a core germplasm collection using microsatellite markers. Ind. Crops Prod. 2020, 145, 112–157. [Google Scholar] [CrossRef]
- Zhang, N.; Wu, W.; Li, S.; Wang, Y.; Ma, Y.; Meng, X.; Zhang, Y. Comprehensive Evaluation of Paddy Quality by Different Drying Methods, Based on Gray Relational Analysis. Agriculture 2022, 12, 1857–1870. [Google Scholar] [CrossRef]
- Zheng, P.; Zhang, M.; Wang, Z.; Wang, T.; Tang, L.; Ma, E.; Liu, J.; Shi, F. Comprehensive evaluation of the fruit quality of the main cultivars of pear (Pyrus spp.) in north China. Erwerbs-Obstbau 2022, 64, 219–227. [Google Scholar] [CrossRef]
- Han, W.; Yang, Z.; Huang, L.; Sun, C.; Yu, X.; Zhao, M. Fuzzy comprehensive evaluation of the effects of relative air humidity on the morpho-physiological traits of Pakchoi (Brassica chinensis L.) under high temperature. Sci. Hortic. 2019, 246, 971–978. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Liu, K.; Muse, S.V. PowerMarker: An integrated analysis environment for genetic marker analysis. Bioinformatics 2005, 21, 2128–2129. [Google Scholar] [CrossRef] [Green Version]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- De Beukelaer, H.; Davenport, G.F.; Fack, V. Core Hunter 3: Flexible core subset selection. BMC Bioinform. 2018, 19, 203–215. [Google Scholar] [CrossRef]
- The SPSSAU Project. SPSSAU (Version 23.0) [Online Application Software]. Available online: https://www.spssau.com (accessed on 13 March 2023).
- Kozak, M.; Kang, M.S. R: Modern Tool for Scientific Computing. Nat. Sci. 2007, 5, 41–43. [Google Scholar]
- Chen, C. Extensions of the TOPSIS for group decision-making under fuzzy environment. Fuzzy Sets Syst. 2000, 114, 1–9. [Google Scholar] [CrossRef]
- Gong, F.; Geng, Y.; Zhang, P.; Zhang, F.; Fan, X.; Liu, Y. Genetic diversity and structure of a core collection of Huangqi (Astragalus ssp.) developed using genomic simple sequence repeat markers. Genet. Resour. Crop Evol. 2022, 70, 571–585. [Google Scholar] [CrossRef]
- Cuevas, H.E.; Prom, L.K. Evaluation of genetic diversity, agronomic traits, and anthracnose resistance in the NPGS Sudan Sorghum Core collection. Bmc Genom. 2020, 21, 88–103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soleimani, B.; Lehnert, H.; Keilwagen, J.; Plieske, J.; Ordon, F.; Naseri Rad, S.; Ganal, M.; Beier, S.; Perovic, D. Comparison between core set selection methods using different Illumina marker platforms: A case study of assessment of diversity in wheat. Front. Plant Sci. 2020, 11, 1040–1051. [Google Scholar] [CrossRef] [PubMed]
- Kelblerová, R.; Dvořák, J.; Korecký, J. Genetic Diversity Maximization as a Strategy for Resilient Forest Ecosystems: A Case Study on Norway Spruce. Forests 2022, 13, 489–502. [Google Scholar] [CrossRef]
- Nie, X.; Wang, Z.; Liu, N.; Li, S.; Yan, B.; Yu, X.; Zhang, Q.; Fang, K.; Zhao, Y.; Xin, C. Fingerprinting 146 Chinese chestnut (Castanea mollissima Blume) accessions and selecting a core collection using SSR markers. J. Integr. Agric. 2021, 20, 1277–1286. [Google Scholar] [CrossRef]
- Razieh, M.; Mohammad, R.D.; Darab, H.; Mehrshad, Z.; Elisa, V.; Sabrina, M.; Fariborz, Z.N. Development of a core collection in Iranian walnut (Juglans regia L.) germplasm using the phenotypic diversity. Sci. Hortic. 2019, 249, 439–448. [Google Scholar]
- Dong, W.; Tian, H.; Wang, D.; Yu, L.; Duan, X.; Liu, S.; Chen, D. Development of 28 EST-SSR markers based on transcriptome sequences of Gobiobotia filifer and cross-species amplification. J. Appl. Ichthyol. 2019, 35, 1295–1299. [Google Scholar] [CrossRef]
- Brown, A.H.D. Core collections: A practical approach to genetic resources management. Genome 1989, 31, 818–824. [Google Scholar] [CrossRef]
- Duan, H.; Cao, S.; Zheng, H.; Hu, D.; Lin, J.; Cui, B.; Lin, H.; Hu, R.; Wu, B.; Sun, Y.; et al. Genetic Characterization of Chinese fir from Six Provinces in Southern China and Construction of a Core Collection. Sci. Rep. 2017, 7, 138–148. [Google Scholar] [CrossRef] [Green Version]
- Yanfang, Z.; Dechang, H.; Jincheng, Z.; Ping, Z.; Zhaohong, W.; Chuanjie, C. Development of a mulberry core collection originated in China to enhance germplasm conservation. Crop Breed. Appl. Biotechnol. 2019, 19, 55–61. [Google Scholar] [CrossRef]
- Sa, K.J.; Kim, D.M.; Oh, J.S.; Park, H.; Hyun, D.Y.; Lee, S.; Rhee, J.H.; Lee, J.K. Construction of a core collection of native Perilla germplasm collected from South Korea based on SSR markers and morphological characteristics. Sci. Rep. 2021, 11, 238–251. [Google Scholar] [CrossRef]
- Xue, H.; Yu, X.; Fu, P.; Liu, B.; Zhang, S.; Li, J.; Zhai, W.; Lu, N.; Zhao, X.; Wang, J.; et al. Construction of the Core Collection of Catalpa fargesii f. duclouxii (Huangxinzimu) Based on Molecular Markers and Phenotypic Traits. Forests 2021, 12, 1518–1529. [Google Scholar] [CrossRef]
- Liu, J.; Xiaomin, L.; Yueqin, L.; Chengyu, X.; Ying, X.; Guolin, C.; Jian, L. Evaluation of genetic diversity and development of core collections of industrial brewing yeast using ISSR markers. Arch. Microbiol. 2020, 203, 1–8. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, T.; Xue, W.; Sun, X.; Liu, H.; Wang, D.; Wang, H.; Wang, X.; Zhang, G.; Wei, Z. Develop a preliminary core germplasm with the novel polymorphism EST-SSRs derived from three transcriptomes of colored calla lily (Zantedeschia hybrida). Front. Plant Sci. 2023, 14, 73–85. [Google Scholar] [CrossRef]
- Yang, H.; Liu, Q.; Zhang, R.; Zhou, Z. Genetic diversity of second generation-parental germplasm of masson pine revealed by SSR markers and establishment of a core germplasm collection. Scand. J. For. Res. 2021, 36, 524–531. [Google Scholar] [CrossRef]
- Bhattacharjee, R.; Khairwal, I.S.; Bramel, P.J.; Reddy, K.N. Establishment of a pearl millet [Pennisetum glaucum (L.) R. Br.] core collection based on geographical distribution and quantitative traits. Euphytica 2007, 155, 35–45. [Google Scholar] [CrossRef] [Green Version]
- Upadhyaya, H.D.; Ortiz, R.; Bramel, P.J.; Singh, S. Development of a groundnut core collection using taxonomical, geographical and morphological descriptors. Genet. Resour. Crop Evol. 2003, 50, 139–148. [Google Scholar] [CrossRef]
- Li, Y.; Shi, Y.; Cao, Y.; Wang, T. Establishment of a core collection for maize germplasm preserved in Chinese National Genebank using geographic distribution and characterization data. Genet. Resour. Crop Evol. 2005, 51, 845–852. [Google Scholar] [CrossRef]
- Giauque, H.; Connor, E.W.; Hawkes, C.V. Endophyte traits relevant to stress tolerance, resource use and habitat of origin predict effects on host plants. New Phytol. 2019, 221, 2239–2249. [Google Scholar] [CrossRef]
- Jin, N.; Jin, L.; Luo, S.; Tang, Z.; Liu, Z.; Wei, S.; Liu, F.; Zhao, X.; Yu, J.; Zhong, Y. Comprehensive evaluation of amino acids and polyphenols in 69 varieties of green cabbage (Brassica oleracea L. var. capitata L.) based on multivariate statistical analysis. Molecules 2021, 26, 5355–5370. [Google Scholar] [CrossRef]
- Laughlin, D.C.; Gremer, J.R.; Adler, P.B.; Mitchell, R.M.; Moore, M.M. The net effect of functional traits on fitness. Trends Ecol. Evol. 2020, 35, 1037–1047. [Google Scholar] [CrossRef]
- Dargan, S.; Kumar, M. A comprehensive survey on the biometric recognition systems based on physiological and behavioral modalities. Expert Syst. Appl. 2020, 143, 113–140. [Google Scholar] [CrossRef]
- Cao, X.; Jiang, F.; Wang, X.; Zang, Y.; Wu, Z. Comprehensive evaluation and screening for chilling-tolerance in tomato lines at the seedling stage. Euphytica 2015, 205, 569–584. [Google Scholar] [CrossRef]
- Liu, X.; Wang, S.; Deng, X.; Zhang, Z.; Yin, L. Comprehensive evaluation of physiological traits under nitrogen stress and participation of linolenic acid in nitrogen-deficiency response in wheat seedlings. BMC Plant Biol. 2020, 20, 501–517. [Google Scholar] [CrossRef] [PubMed]
- Şahin, C.B.; İşler, N. Foliar applied zinc and iron effects on yield and yield components of soybean: Determination by PCA analysis. Commun. Soil Sci. Plant Anal. 2021, 52, 212–221. [Google Scholar] [CrossRef]
- Verma, M.K.; Lal, S.; Sharma, V.K.; Choudhary, H.; Ahmed, N. PCA and genetic divergence analysis in pomegranate (Punica granatum L.) cultivars. Int. J. Innov. Hortic. 2019, 8, 45–50. [Google Scholar]
- Wang, B.; Xie, H.; Ren, H.; Li, X.; Chen, L.; Wu, B. Application of AHP, TOPSIS, and TFNs to plant selection for phytoremediation of petroleum-contaminated soils in shale gas and oil fields. J. Clean. Prod. 2019, 233, 13–22. [Google Scholar] [CrossRef]
- Wang, Z.; Yang, P.; Peng, H.; Li, C.; Yue, C.; Li, W.; Jiang, X. Comprehensive evaluation of 47 tea [Camellia sinensis (L.) O. Kuntze] germplasm based on entropy weight method and grey relational degree. Genet. Resour. Crop Evol. 2021, 68, 3257–3270. [Google Scholar] [CrossRef]
- Çelikbilek, Y.; Tüysüz, F. An in-depth review of theory of the TOPSIS method: An experimental analysis. J. Manag. Anal. 2020, 7, 281–300. [Google Scholar] [CrossRef]
- De Lima Silva, D.F.; De Almeida Filho, A.T. Sorting with TOPSIS through boundary and characteristic profiles. Comput. Ind. Eng. 2020, 141, 1063–1078. [Google Scholar] [CrossRef]
- Li, Z.; Luo, Z.; Wang, Y.; Fan, G.; Zhang, J. Suitability evaluation system for the shallow geothermal energy implementation in region by Entropy Weight Method and TOPSIS method. Renew. Energy 2022, 184, 564–576. [Google Scholar] [CrossRef]
- Liang, C.; Yu, S.; Zhang, H.; Wang, Z.; Li, F. Economic Evaluation of Drought Resistance Measures for Maize Seed Production Based on TOPSIS Model and Combination Weighting Optimization. Water 2022, 14, 3262–3280. [Google Scholar] [CrossRef]
- Chen, P. Effects of the entropy weight on TOPSIS. Expert Syst. Appl. 2020, 168, 114–186. [Google Scholar] [CrossRef]
- Abdi, H.; Williams, L.J. Principal component analysis. Wiley Interdiscip. Rev. Comput. Stat. 2010, 2, 433–459. [Google Scholar] [CrossRef]
- Makowski, D.; Ben-Shachar, M.S.; Patil, I.; Lüdecke, D. Methods and algorithms for correlation analysis in R. J. Open Source Softw. 2020, 5, 2306–2310. [Google Scholar] [CrossRef]


| Loci | Motifs | Target _Length (bp) | Primer_Sequence |
|---|---|---|---|
| TC10 | TC (7) | 182 | F: TAGAGACAAGTTTGAGTGGAGCG R: GCATGTGATGTAGGAGTCTGAACA |
| TC11 | GA (8) | 232 | F: ACCATGTCAAGAAACCTTTTGTAACA R: TGAGGCTAAATGTGCATCTCTTGA |
| TcB27 | AG (8) | 191 | F: GGCAGAGAAGAGCGGTTTTA R: CGGATCTTTCGCAACGTAGT |
| XC107 | CA (19) | 238 | F: GGAATTAATCAAGGTTACGCATGCA R: ACTCTTTCCCTAACTTATGGTGATTTCA |
| XC193 | TG (16) | 237 | F: TGAATGTGGCTAGTCTGGAAAATTT R: TCTCTTAAGCCTCGATGATGTGT |
| XC227 | TC (17) | 263 | F: AGATGCCTTCTTGAGCTTGAAAGA R: GGTTATTCCCAAGGTCAACAGAAA |
| XC239 | CA (14) | 277 | F: ACATAACAACCGTCACACACTCG R: CAGTCCACACCCCAAACTTAGAT |
| XC301 | TG (25) | 242 | F: CCCACCGACCTCACTTTAAATCT R: TCCAACACAATCACGTCATTCTCA |
| XC316 | AG (23) | 248 | F: TCCAAGAGAAATCCACCACTTGA R: TGACCATTCTACCCTTATGTTCAGA |
| XC320 | AG (15) | 256 | F: GGCCACTCCTGCATACACAA R: AGACATGGTGGCCCTCCTAC |
| XC35 | CT (10) | 259 | F: TGACATGATGGCGATTTACAGGT R: TGTTAAACCTTCTCCTGACTAATCCA |
| XC41 | AC (12) | 186 | F: GCTTTACTGGGATTGCTGGGAAT R: TTTACACTGAACTCTGCAATCACTT |
| XC66 | CAT (9) | 190 | F: TATGGCCCATGATCATCGTCAAC R: AGTGTGATGTAGAGGAGGTGGAG |
| Loci | Na | Ne | I | Ho | He | PIC |
|---|---|---|---|---|---|---|
| TC10 | 27 | 9.922 | 2.7 | 0.639 | 0.899 | 0.893 |
| TC11 | 20 | 3.869 | 1.572 | 0.481 | 0.742 | 0.697 |
| TcB27 | 15 | 4.138 | 1.63 | 0.643 | 0.758 | 0.721 |
| XC107 | 35 | 13.137 | 2.906 | 0.506 | 0.924 | 0.919 |
| XC193 | 26 | 11.259 | 2.756 | 0.513 | 0.911 | 0.906 |
| XC227 | 22 | 11.281 | 2.608 | 0.811 | 0.911 | 0.905 |
| XC239 | 19 | 2.234 | 1.482 | 0.524 | 0.552 | 0.542 |
| XC301 | 41 | 9.807 | 2.807 | 0.448 | 0.898 | 0.892 |
| XC316 | 30 | 18.419 | 3.084 | 0.603 | 0.946 | 0.943 |
| XC320 | 23 | 5.995 | 2.282 | 0.942 | 0.833 | 0.822 |
| XC35 | 15 | 5.284 | 1.976 | 0.696 | 0.811 | 0.792 |
| XC41 | 20 | 7.263 | 2.319 | 0.779 | 0.862 | 0.851 |
| XC66 | 9 | 2.879 | 1.292 | 0.498 | 0.653 | 0.59 |
| Mean | 23.231 | 8.114 | 2.263 | 0.622 | 0.823 | 0.806 |
| Cluster | Number | Proportion | |
|---|---|---|---|
| Cluster1 | 263 | 25.29% | |
| Core | 23 | 2.21% | |
| Retain | 240 | 23.08% | |
| Cluster2 | 777 | 74.71% | |
| Core | 185 | 17.79% | |
| Retain | 592 | 56.92% | |
| Total | 1040 | 100.00% |
| Traits | Min | Max | Mean ± SD | CV |
|---|---|---|---|---|
| H/cm | 40.00 | 280.00 | 160.09 ± 45.058 | 28.15% |
| FZS/unit | 1.00 | 9.00 | 3.04 ± 1.584 | 52.15% |
| ZCFY/cm | 25.00 | 102.67 | 63.50 ± 11.866 | 18.69% |
| FYDS/pair | 8.00 | 24.50 | 15.25 ± 3.260 | 21.38% |
| FYSL/unit | 10.00 | 70.00 | 33.03 ± 10.879 | 32.93% |
| YBM/grade | 1.00 | 5.00 | 2.04 ± 1.309 | 64.06% |
| FYTL/unit | 0.00 | 9.00 | 2.31 ± 2.127 | 92.16% |
| FYBS/unit | 0.00 | 34.00 | 4.82 ± 3.713 | 77.07% |
| Traits | PCA1 | PCA2 | PCA3 | Comprehensive Score | Weight |
|---|---|---|---|---|---|
| H | 0.412 | 0.482 | 0.138 | 0.377 | 15.42% |
| FZS | 0.078 | 0.080 | 0.696 | 0.219 | 8.97% |
| ZCFY | 0.574 | 0.006 | 0.290 | 0.293 | 12.01% |
| FYDS | 0.479 | 0.299 | 0.284 | 0.366 | 15.00% |
| FYSL | 0.265 | 0.543 | 0.160 | 0.347 | 14.20% |
| YBM | 0.420 | 0.317 | 0.182 | 0.327 | 13.37% |
| FYTL | 0.112 | 0.282 | 0.462 | 0.256 | 10.49% |
| FYBS | 0.084 | 0.445 | 0.243 | 0.257 | 10.53% |
| Eigenvalue | 1.958 | 1.899 | 1.132 | ||
| Variance interpretation rate | 24.47% | 23.73% | 14.15% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dai, J.; Fan, Y.; Diao, S.; Yin, H.; Han, X.; Liu, J. Construction of Core Collection and Phenotypic Evaluation of Toona sinensis. Forests 2023, 14, 1269. https://doi.org/10.3390/f14061269
Dai J, Fan Y, Diao S, Yin H, Han X, Liu J. Construction of Core Collection and Phenotypic Evaluation of Toona sinensis. Forests. 2023; 14(6):1269. https://doi.org/10.3390/f14061269
Chicago/Turabian StyleDai, Jianhua, Yanru Fan, Shu Diao, Hengfu Yin, Xiaojiao Han, and Jun Liu. 2023. "Construction of Core Collection and Phenotypic Evaluation of Toona sinensis" Forests 14, no. 6: 1269. https://doi.org/10.3390/f14061269






