Influence of Spatiotemporal Dynamics on the Fine-Scale Spatial Genetic Structure of Differently Managed Picea abies Stands
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. Sampling Strategy and Genotyping
2.3. Spatial Data Analysis
2.4. Genetic Data Analysis
2.5. Relationship between Age and Fine-Scale SGS
3. Results
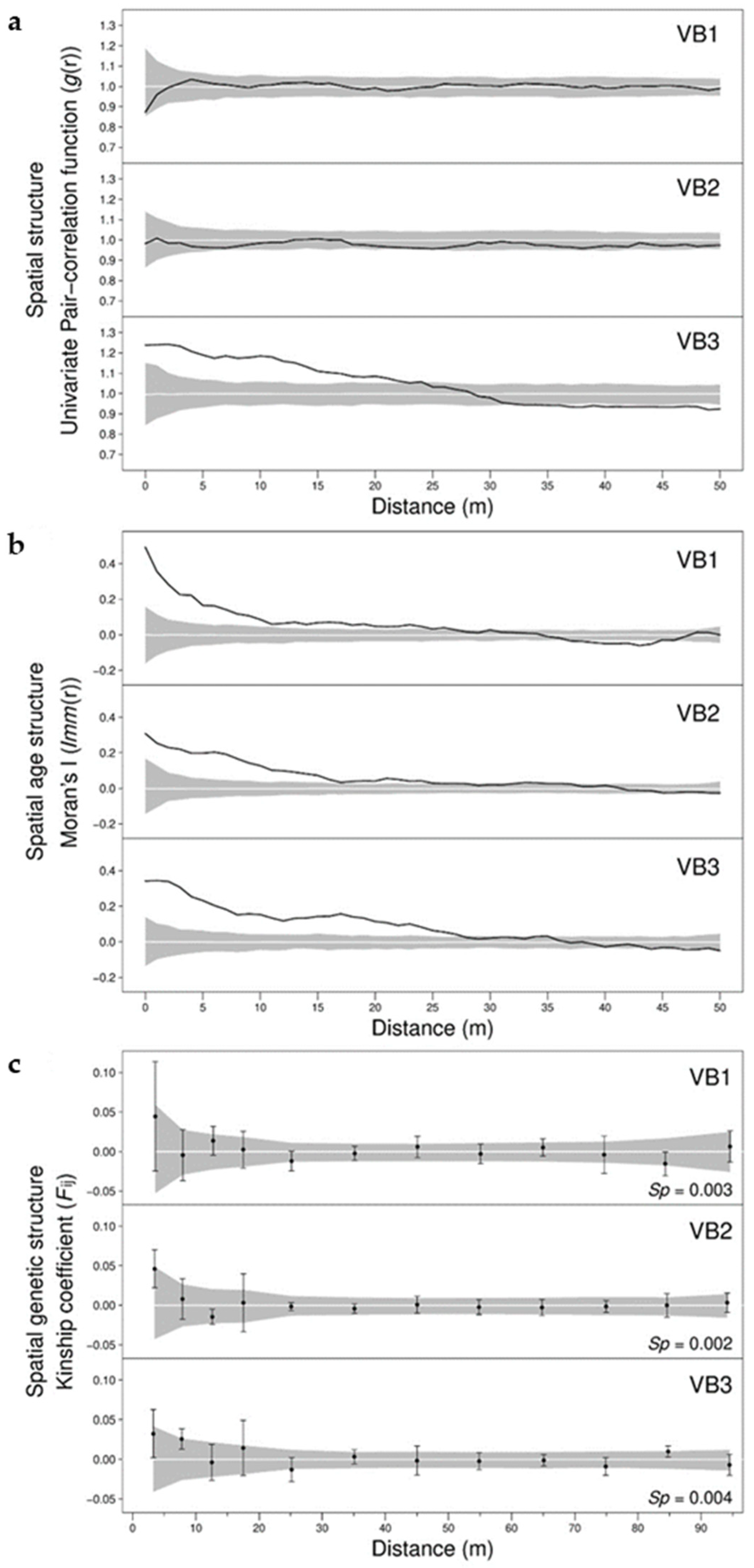
3.1. Spatial Distribution of Individual Age
3.2. Spatial Distribution of Genetic Variation
3.3. Relationship between Age and Fine-Scale SGS
4. Discussion
4.1. Influence of Forest Management on the Spatial and Age Structure
4.2. Influence of Forest Management on the Fine-Scale Spatial Genetic Structure
4.3. Effect of Spatiotemporal Dynamics on the Fine-Scale Spatial Genetic Structure
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Franklin, J.F.; Lindenmayer, D.; Thornburgh, D.; van Pelt, R.; Chen, J.; Spies, T.A.; Carey, A.B.; Shaw, D.C.; Berg, D.R.; Harmon, M.E.; et al. Disturbances and structural development of natural forest ecosystems with silvicultural implications, using Douglas-fir forests as an example. For. Ecol. Manag. 2002, 155, 399–423. [Google Scholar] [CrossRef]
- Motta, R.; Garbarino, M.; Berretti, R.; Meloni, F.; Nosenzo, A.; Vacchiano, G. Development of old-growth characteristics in uneven-aged forests of the Italian Alps. Eur. J. For. Res. 2015, 134, 19–31. [Google Scholar] [CrossRef]
- Moeur, M. Characterizing spatial patterns of trees using stem-mapped data. For. Sci. 1993, 39, 756–775. [Google Scholar]
- Adams, W.T.; Zuo, J.; Shimizu, J.Y.; Tappeiner, J.C. Impact of alternative regeneration methods on genetic diversity in coastal Douglas-fir. For. Sci. 1998, 44, 390–396. [Google Scholar]
- Rajora, O.P. Genetic biodiversity impacts of silvicultural practices and phenotypic selection in white spruce. Theor. Appl. Genet. 1999, 99, 954–961. [Google Scholar] [CrossRef]
- Epperson, B.K.; Chung, M.G. Spatial genetic structure of allozyme polymorphisms within populations of Pinus strobus (Pinaceae). Am. J. Bot. 2001, 88, 1006–1010. [Google Scholar] [CrossRef] [PubMed]
- Marquardt, P.E.; Epperson, B.K. Spatial and population genetic structure of microsatellites in white pine. Mol. Ecol. 2004, 13, 3305–3315. [Google Scholar] [CrossRef] [PubMed]
- Marquardt, P.E.; Echt, C.S.; Epperson, B.K.; Pubanz, D.M. Genetic structure, diversity, and inbreeding of eastern white pine under different management conditions. Can. J. For. Res. 2007, 37, 2652–2662. [Google Scholar] [CrossRef]
- Kavaliauskas, D.; Fussi, B.; Westergren, M.; Aravanopoulos, F.; Finzgar, D.; Baier, R.; Alizoti, P.; Bozic, G.; Avramidou, E.; Konnert, M.; et al. The interplay between forest management practices, genetic monitoring, and other long-term monitoring systems. Forests 2018, 9, 133. [Google Scholar] [CrossRef]
- Neale, D.B. Genetic implications of shelterwood regeneration of Douglas-fir in Southwest Oregon. For. Sci. 1985, 15, 995–1005. [Google Scholar]
- Piotti, A.; Leonardi, S.; Heuertz, M.; Buiteveld, J.; Geburek, T.; Gerber, S.; Kramer, K.; Vettori, C.; Vendramin, G.G. Within-population genetic structure in beech (Fagus sylvatica L.) stands characterized by different disturbance histories: Does forest management simplify population substructure? PLoS ONE 2013, 8, e73391. [Google Scholar] [CrossRef] [PubMed]
- Piotti, A. The genetic consequences of habitat fragmentation: The case of forests. iForest—Biogeosci. For. 2009, 2, 75–76. [Google Scholar] [CrossRef]
- Lowe, A.J.; Cavers, S.; Boshier, D.; Breed, M.F.; Hollingsworth, P.M. The resilience of forest fragmentation genetics—No longer a paradox—We were just looking in the wrong place. Heredity 2015, 115, 97–99. [Google Scholar] [CrossRef] [PubMed]
- Rajendra, K.C.; Seifert, S.; Prinz, K.; Gailing, O.; Finkeldey, R. Subtle human impacts on neutral genetic diversity and spatial patterns of genetic variation in European beech (Fagus sylvatica). For. Ecol. Manag. 2014, 319, 138–149. [Google Scholar] [CrossRef]
- Sjölund, M.J.; Jump, A.S. Coppice management of forests impacts spatial genetic structure but not genetic diversity in European beech (Fagus sylvatica L.). For. Ecol. Manag. 2015, 336, 65–71. [Google Scholar] [CrossRef]
- Lamedica, S.; Lingua, E.; Popa, I.; Motta, R.; Carrer, M. Spatial structure in four Norway spruce stands with different management history in the Alps and Carpathians. Silva Fenn. 2011, 45, 865–873. [Google Scholar] [CrossRef]
- Westergren, M.; Bozic, G.; Ferreira, A.; Kraigher, H. Insignificant effect of management using irregular shelterwood system on the genetic diversity of European beech (Fagus sylvatica L.): A case study of managed stand and old growth forest in Slovenia. For. Ecol. Manag. 2015, 335, 51–59. [Google Scholar] [CrossRef]
- Bontemps, A.; Klein, E.K.; Oddou-Muratorio, S. Shift of spatial patterns during early recruitment in Fagus sylvatica: Evidence from seed dispersal estimates based on genotypic data. For. Ecol. Manag. 2013, 305, 67–76. [Google Scholar] [CrossRef]
- Troupin, D.; Nathan, R.; Vendramin, G.G. Analysis of spatial genetic structure in an expanding Pinus halepensis population reveals development of fine-scale genetic clustering over time. Mol. Ecol. 2006, 15, 3617–3630. [Google Scholar] [CrossRef] [PubMed]
- Lesser, M.R.; Parchman, T.L.; Jackson, S.T. Development of genetic diversity, differentiation and structure over 500 years in four ponderosa pine populations. Mol. Ecol. 2013, 22, 2640–2652. [Google Scholar] [CrossRef] [PubMed]
- King, G.M.; Gugerli, F.; Fonti, P.; Frank, D.C. Tree growth response along an elevational gradient: Climate or genetics? Oecologia 2013, 173, 1587–1600. [Google Scholar] [CrossRef] [PubMed]
- Bosela, M.; Popa, I.; Gömöry, D.; Longauer, R.; Tobin, B.; Kyncl, J.; Kyncl, T.; Nechita, C.; Petráš, R.; Sidor, C.G.; et al. Effects of post-glacial phylogeny and genetic diversity on the growth variability and climate sensitivity of European silver fir. J. Ecol. 2016, 104, 716–724. [Google Scholar] [CrossRef]
- Heer, K.; Behringer, D.; Piermattei, A.; Bässler, C.; Brandl, R.; Fady, B.; Jehl, H.; Liepelt, S.; Lorch, S.; Piotti, A.; et al. Linking dendroecology and association genetics in natural populations: Stress responses archived in tree rings associate with SNP genotypes in silver fir (Abies alba Mill.). Mol. Ecol. 2018, 27, 1428–1438. [Google Scholar] [CrossRef] [PubMed]
- Avanzi, C.; Piermattei, A.; Piotti, A.; Büntgen, U.; Heer, K.; Opgenoorth, L.; Spanu, I.; Urbinati, C.; Vendramin, G.G.; Leonardi, S. Disentangling the effects of spatial proximity and genetic similarity on individual growth performances in Norway spruce natural populations. Sci. Total Environ. 2019, 650, 493–504. [Google Scholar] [CrossRef] [PubMed]
- Lorimer, C.G.; Frelich, L.E. A methodology for estimating canopy disturbance frequency and intensity in dense temperate forests. Can. J. For. Res. 1989, 19, 651–663. [Google Scholar] [CrossRef]
- Carrer, M.; Urbinati, C. Age-dependent tree-ring growth responses to climate in Larix decidua and Pinus cembra. Ecology 2004, 85, 730–740. [Google Scholar] [CrossRef]
- Primicia, I.; Camarero, J.J.; Janda, P.; Čada, V.; Morrissey, R.C.; Trotsiuk, V.; Bače, R.; Teodosiu, M.; Svoboda, M. Age, competition, disturbance and elevation effects on tree and stand growth response of primary Picea abies forest to climate. For. Ecol. Manag. 2015, 354, 77–86. [Google Scholar] [CrossRef]
- Rita, A.; Borghetti, M.; Todaro, L.; Saracino, A. Interpreting the Climatic Effects on Xylem Functional Traits in Two Mediterranean Oak Species: The Role of Extreme Climatic Events. Front. Plant Sci. 2016, 7, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Motta, R.; Nola, P.; Piussi, P. Structure and stand development in three subalpine Norway spruce (Picea abies (L.) Karst.) stands in Paneveggio (Trento, Italy). Glob. Ecol. Biogeogr. 1999, 8, 455–471. [Google Scholar] [CrossRef]
- Duncker, P.S.; Barreiro, S.M.; Hengeveld, G.M.; Lind, T.; Mason, W.L.; Ambrozy, S. Classification of Forest Management Approaches: A New Conceptual Framework and Its Applicability to European Forestry. Ecol. Soc. 2012, 17, 51. [Google Scholar] [CrossRef]
- Altman, J.; Hédl, R.; Szabó, P.; Mazůrek, P.; Riedl, V.; Müllerová, J.; Kopecký, M.; Doležal, J. Tree-rings mirror management legacy: Dramatic response of standard oaks to past coppicing in Central Europe. PLoS ONE 2013, 8, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Austerlitz, F.; Mariette, S.; Machon, N.; Gouyon, P.H.; Godelle, B. Effects of colonization process on genetic diversity: Differences between annual plant and tree species. Genetics 2000, 154, 1309–1321. [Google Scholar] [PubMed]
- Motta, R. Old-growth forests and silviculture in the Italian Alps: The case-study of the strict reserve of Paneveggio (TN). Plant Biosyst. 2002, 136, 223–231. [Google Scholar] [CrossRef]
- Motta, R.; Nola, P.; Piussi, P. Long-term investigations in a strict forest reserve in the eastern Italian Alps: Spatio-temporal origin and development in two multi-layered subalpine stands. J. Ecol. 2002, 90, 495–507. [Google Scholar] [CrossRef]
- Di Tommaso, P.L. Contributo ad una tipologia floristicoecologica della foresta di Paneveggio (Trento). Versante meridionale. Ann. Accad. Ital. Sci. For. 1983, 32, 287–315. [Google Scholar]
- Motta, R.; Nola, P. Fraying damages in the subalpine forest of Paneveggio (Trento, Italy): A dendroecological approach. For. Ecol. Manag. 1996, 88, 81–86. [Google Scholar] [CrossRef]
- Scotti, I.; Paglia, G.P.; Magni, F.; Morgante, M. Efficient development of dinucleotide microsatellite markers in Norway spruce (Picea abies Karst.) through dot-blot selection. Theor. Appl. Genet. 2002, 104, 1035–1041. [Google Scholar] [CrossRef] [PubMed]
- Pfeiffer, A.; Olivieri, A.; Morgante, M. Identification and characterization of microsatellites in Norway spruce (Picea abies K.). Genome 1997, 40, 411–419. [Google Scholar] [CrossRef] [PubMed]
- Piotti, A.; Leonardi, S.; Piovani, P.; Scalfi, M.; Menozzi, P. Spruce colonization at treeline: Where do those seeds come from. Heredity 2009, 103, 136–145. [Google Scholar] [CrossRef] [PubMed]
- Shimatani, K. Point processes for fine-scale spatial genetics and molecular ecology. Biom. J. 2002, 44, 325–352. [Google Scholar] [CrossRef]
- Wiegand, T.; Moloney, K.A. Handbook of Spatial Point-pattern Analysis in Ecology; Chapman and Hall/CRC: New York, NY, USA, 2014. [Google Scholar]
- Stoyan, D.; Stoyan, H. Fractals, Random Shapes, and Point Fields: Methods of Geometrical Statistics; Wiley: Chichester, UK, 1994. [Google Scholar]
- Wiegand, T.; Moloney, K.A. Rings, circles, and null-models for point pattern analysis in ecology. Oikos 2004, 104, 209–229. [Google Scholar] [CrossRef] [Green Version]
- Diggle, P.J. Statistical Analysis of Spatial Point Patterns; Edward Arnold: London, UK, 2003. [Google Scholar]
- Getis, A.; Ord, J.K. The analysis of spatial association by use of distance statistics. Geogr. Anal. 1992, 24, 189–206. [Google Scholar] [CrossRef]
- Carrer, M.; Soraruf, L.; Lingua, E. Convergent space-time tree regeneration patterns along an elevation gradient at high altitude in the Alps. For. Ecol. Manag. 2013, 304, 1–9. [Google Scholar] [CrossRef]
- Sawada, M. Rookcase: An Excel 97/2000 Visual Basic (VB) add-in for exploring global and local spatial autocorrelation. Bull. Ecol. Soc. Am. 1999, 80, 231–234. [Google Scholar]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [PubMed]
- Kalinowski, S.T. hp-rare 1.0: A computer program for performing rarefaction on measures of allelic richness. Mol. Ecol. Notes 2005, 5, 187–189. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018. [Google Scholar]
- Loiselle, B.A.; Sork, V.L.; Nason, J.; Graham, C. Spatial genetic structure of a tropical understory shrub, Psychotria officinalis (Rubiaceae). Am. J. Bot. 1995, 82, 1420–1425. [Google Scholar] [CrossRef]
- Hardy, O.J.; Vekemans, X. SPAGeDi: A versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol. Ecol. Notes 2002, 2, 618–620. [Google Scholar] [CrossRef]
- Vekemans, X.; Hardy, O.J. New insights from fine-scale spatial genetic structure analyses in plant populations. Mol. Ecol. 2004, 13, 921–935. [Google Scholar] [CrossRef] [PubMed]
- Guillot, G.; Mortier, F.; Estoup, A. Geneland: A computer package for landscape genetics. Mol. Ecol. Notes 2005, 5, 712–715. [Google Scholar] [CrossRef]
- Guillot, G.; Santos, F.; Estoup, A. Analysing georeferenced population genetics data with Geneland: A new algorithm to deal with null alleles and a friendly graphical user interface. Bioinformatics 2008, 24, 1406–1407. [Google Scholar] [CrossRef] [PubMed]
- Schoenenberger, W.; Kuhn, N.; Laessig, R. Research Objectives and Projects on Windthrow Areas in Switzerland; AGRIS: Zurich, Switzerland, 1995. [Google Scholar]
- Motta, R.; Lingua, E. Human impact on size, age, and spatial structure in a mixed European larch and Swiss stone pine forest in the Western Italian Alps. Can. J. For. Res. 2005, 35, 1809–1820. [Google Scholar] [CrossRef]
- Resler, L.M.; Butler, D.R.; Malanson, G.P. Topographic shelter and conifer establishment and mortality in an alpine environment, Glacier National Park, Montana. Phys. Geogr. 2005, 26, 112–125. [Google Scholar] [CrossRef]
- Maher, E.L.; Germino, M.J. Microsite differentiation among conifer species during seedling establishment at alpine treeline. Ecoscience 2006, 13, 334–341. [Google Scholar] [CrossRef] [Green Version]
- Kuuluvainen, T. Gap disturbance, ground microtopography, and the regeneration dynamics of boreal coniferous forests in Finland: A review. Ann. Zool. Fenn. 1994, 31, 35–51. [Google Scholar]
- Holtmeier, F.K. Mountain Timberlines: Ecology, Patchiness, and Dynamics; Springer Science & Business Media: Berlin, Germany, 2009; Volume 36. [Google Scholar]
- Ratnam, W.; Rajora, O.P.; Finkeldey, R.; Aravanopoulos, F.; Bouvet, J.-M.; Vaillancourt, R.E.; Kanashiro, M.; Fady, B.; Tomita, M.; Vinson, C. Genetic effects of forest management practices: Global synthesis and perspectives. For. Ecol. Manag. 2014, 333, 52–65. [Google Scholar] [CrossRef]
- Aravanopoulos, F. Do silviculture and forest management affect the genetic diversity and structure of long-impacted forest tree populations? Forests 2018, 9, 355. [Google Scholar] [CrossRef]
- Motta, R.; Berretti, R.; Castagneri, D.; Lingua, E.; Nola, P.; Vacchiano, G. Stand and coarse woody debris dynamics in subalpine Norway spruce forests withdrawn from regular management. Ann. For. Sci. 2010, 67, 803. [Google Scholar] [CrossRef]
- Maghuly, F.; Pinsker, W.; Praznik, W.; Fluch, S. Genetic diversity in managed subpopulations of Norway spruce [Picea abies (L.) Karst.]. For. Ecol. Manag. 2006, 222, 266–271. [Google Scholar] [CrossRef]
- Unger, G.M.; Konrad, H.; Geburek, T. Does spatial genetic structure increase with altitude? An answer from Picea abies in Tyrol, Austria. Plant Syst. Evol. 2011, 292, 133–141. [Google Scholar] [CrossRef]
- Evans, M.E.K.; Gugger, P.F.; Lynch, A.M.; Guiterman, C.H.; Fowler, J.C.; Klesse, S.; Riordan, E.C. Dendroecology meets genomics in the common garden: New insights into climate adaptation. New Phytol. 2018, 218, 401–403. [Google Scholar] [CrossRef] [PubMed]
- Trujillo-Moya, C.; George, J.-P.; Fluch, S.; Geburek, T.; Grabner, M.; Karanitsch-Ackerl, S.; Konrad, H.; Mayer, K.; Sehr, E.M.; Wischnitzki, E.; et al. Drought sensitivity of Norway spruce at the species’ warmest fringe: Quantitative and molecular analysis reveals high genetic variation among and within provenances. G3: Genes Genomes Genet. 2018, 8, 1225–1245. [Google Scholar] [CrossRef] [PubMed]
- Housset, J.M.; Nadeau, S.; Isabel, N.; Depardieu, C.; Duchesne, I.; Lenz, P.; Girardin, M.P. Tree rings provide a new class of phenotypes for genetic associations that foster insights into adaptation of conifers to climate change. New Phytol. 2018, 218, 630–645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leonarduzzi, C.; Piotti, A.; Spanu, I.; Vendramin, G.G. Effective gene flow in a historically fragmented area at the southern edge of silver fir (Abies alba Mill.) distribution. Tree Genet. Genomes 2016, 12, 95. [Google Scholar] [CrossRef]




| Plot | N Spatial Analysis 1 | N Genetic Analysis | Elevation (m a.s.l.) | Regeneration Density (n ha−1) | Past Management | Last Intervention | Na | HE | Ar100 |
|---|---|---|---|---|---|---|---|---|---|
| VB1 | 479 | 97 | 1695 | 935 | Wood production | 1984 | 10 | 0.64 | 8.87 |
| VB2 | 541 | 114 | 1815 | 30 | Wood production | 1948 | 11.4 | 0.64 | 9.34 |
| VB3 | 452 | 117 | 1865 | 3010 | Wood pasture | 1929 | 10.6 | 0.64 | 9.21 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Piotti, A.; Garbarino, M.; Avanzi, C.; Berretti, R.; Motta, R.; Piovani, P.; Leonardi, S. Influence of Spatiotemporal Dynamics on the Fine-Scale Spatial Genetic Structure of Differently Managed Picea abies Stands. Forests 2018, 9, 622. https://doi.org/10.3390/f9100622
Piotti A, Garbarino M, Avanzi C, Berretti R, Motta R, Piovani P, Leonardi S. Influence of Spatiotemporal Dynamics on the Fine-Scale Spatial Genetic Structure of Differently Managed Picea abies Stands. Forests. 2018; 9(10):622. https://doi.org/10.3390/f9100622
Chicago/Turabian StylePiotti, Andrea, Matteo Garbarino, Camilla Avanzi, Roberta Berretti, Renzo Motta, Paolo Piovani, and Stefano Leonardi. 2018. "Influence of Spatiotemporal Dynamics on the Fine-Scale Spatial Genetic Structure of Differently Managed Picea abies Stands" Forests 9, no. 10: 622. https://doi.org/10.3390/f9100622
APA StylePiotti, A., Garbarino, M., Avanzi, C., Berretti, R., Motta, R., Piovani, P., & Leonardi, S. (2018). Influence of Spatiotemporal Dynamics on the Fine-Scale Spatial Genetic Structure of Differently Managed Picea abies Stands. Forests, 9(10), 622. https://doi.org/10.3390/f9100622





