Chondrocytes Contribute to Alphaviral Disease Pathogenesis as a Source of Virus Replication and Soluble Factor Production
Abstract
:1. Introduction
2. Materials and Methods
2.1. Virus
2.2. Mice and Histology
2.3. Primary Cell Cultures and Infection
2.4. Gene Expression Analysis by qPCR
2.5. Bio-Plex® Multiplex Assay
2.6. Statistical Analysis
3. Results
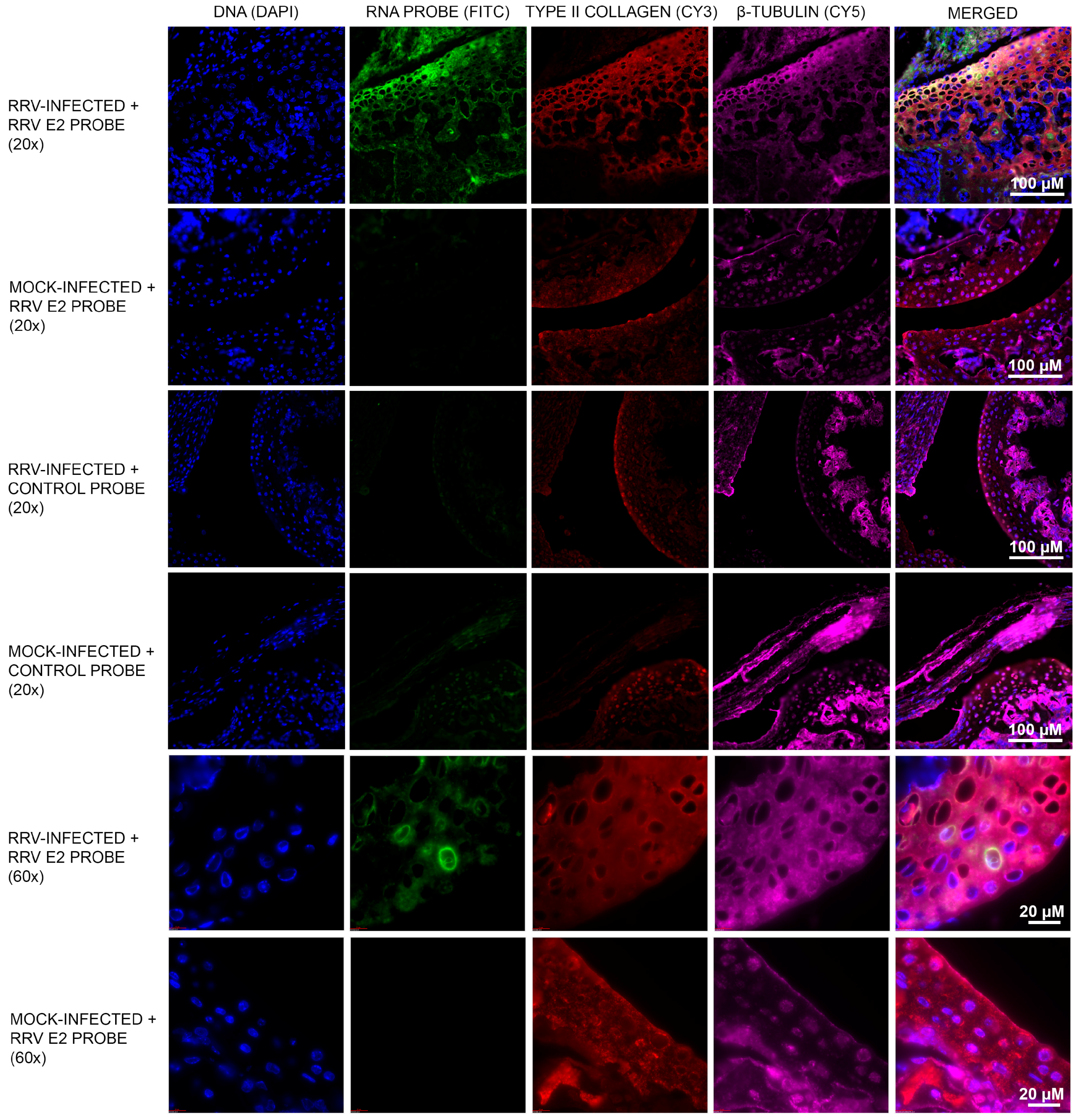
3.1. Murine Chondrocytes Are Susceptible to RRV Infection
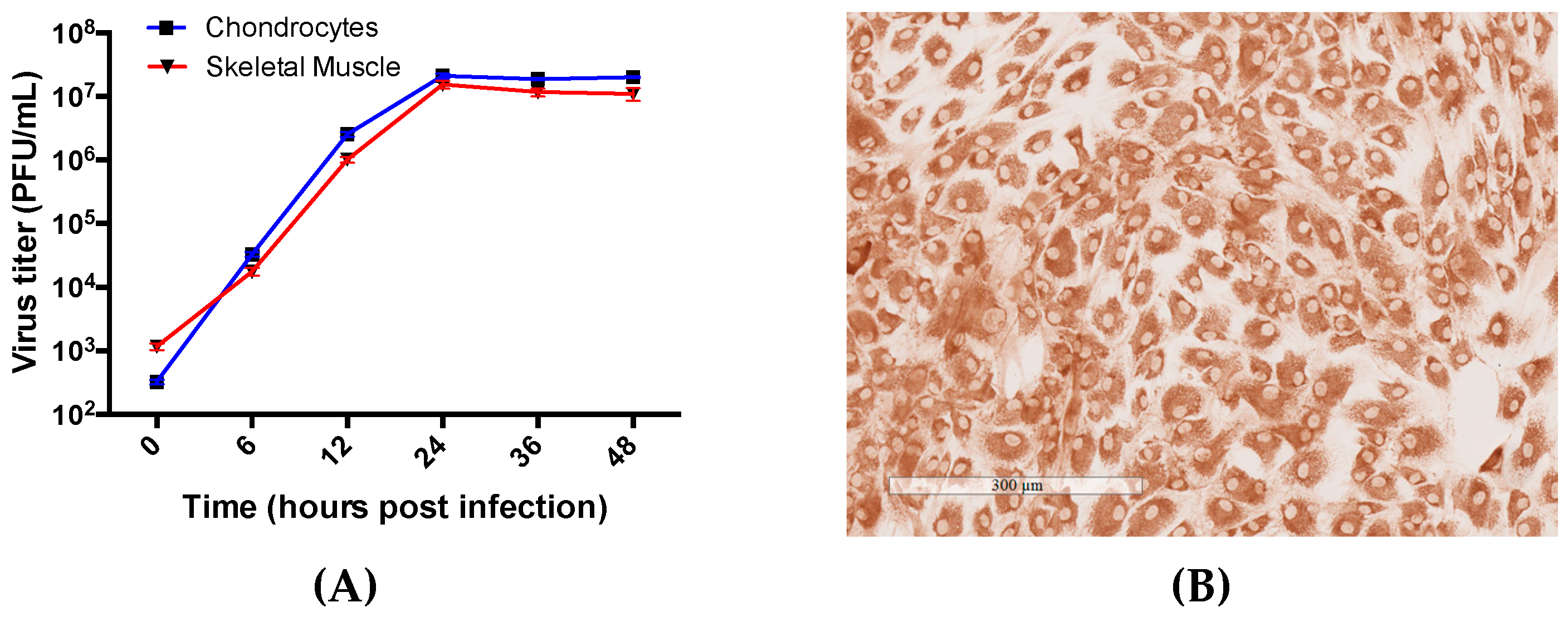
3.2. Primary Human Chondrocyte and Skeletal Muscle Cells Are Permissive to RRV Infection and Supports Productive Virus Replication
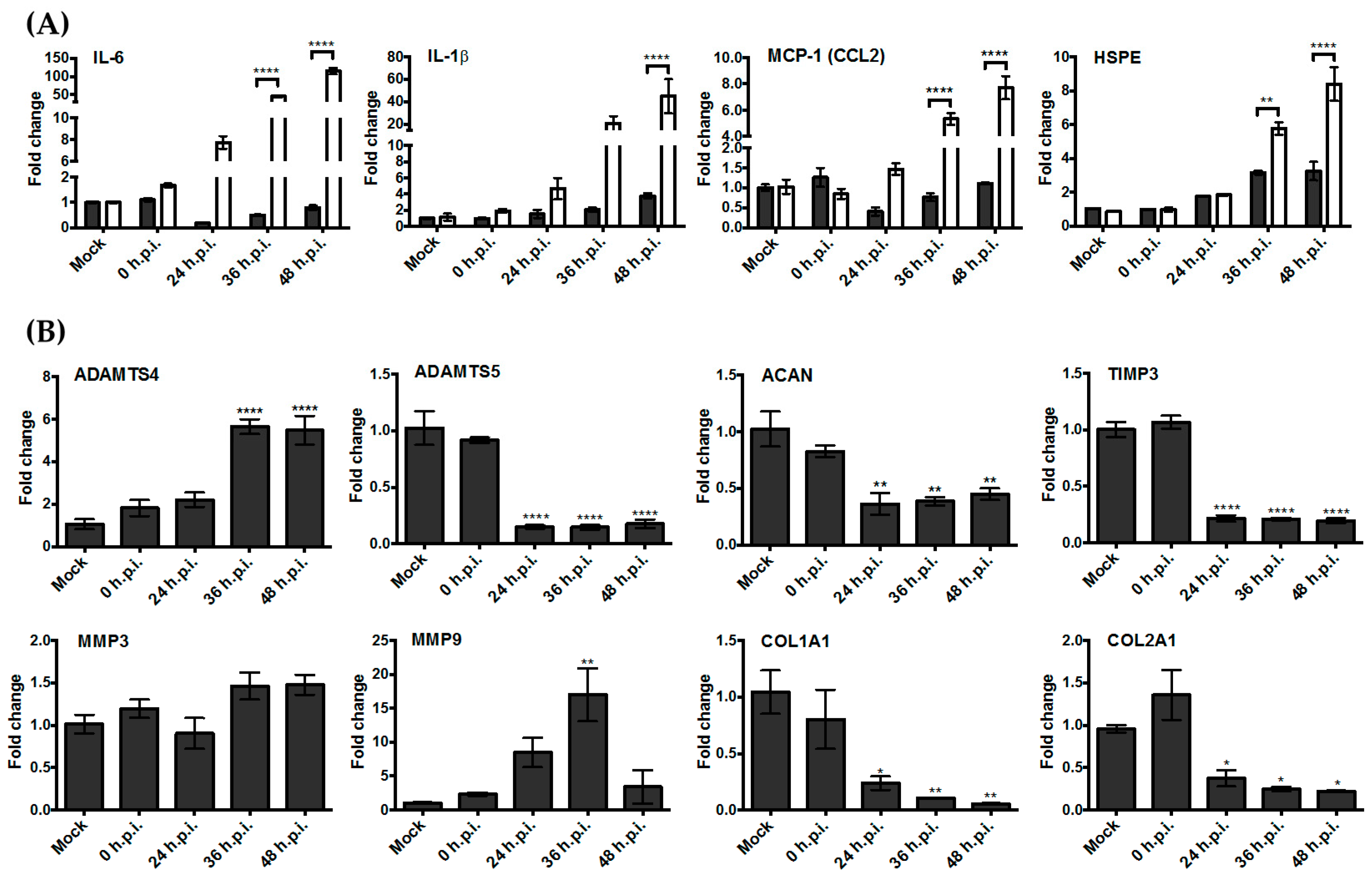
3.3. Upregulation of the Genes Encoding Pro-Inflammatory Factors and Degrading Enzymes Was Observed
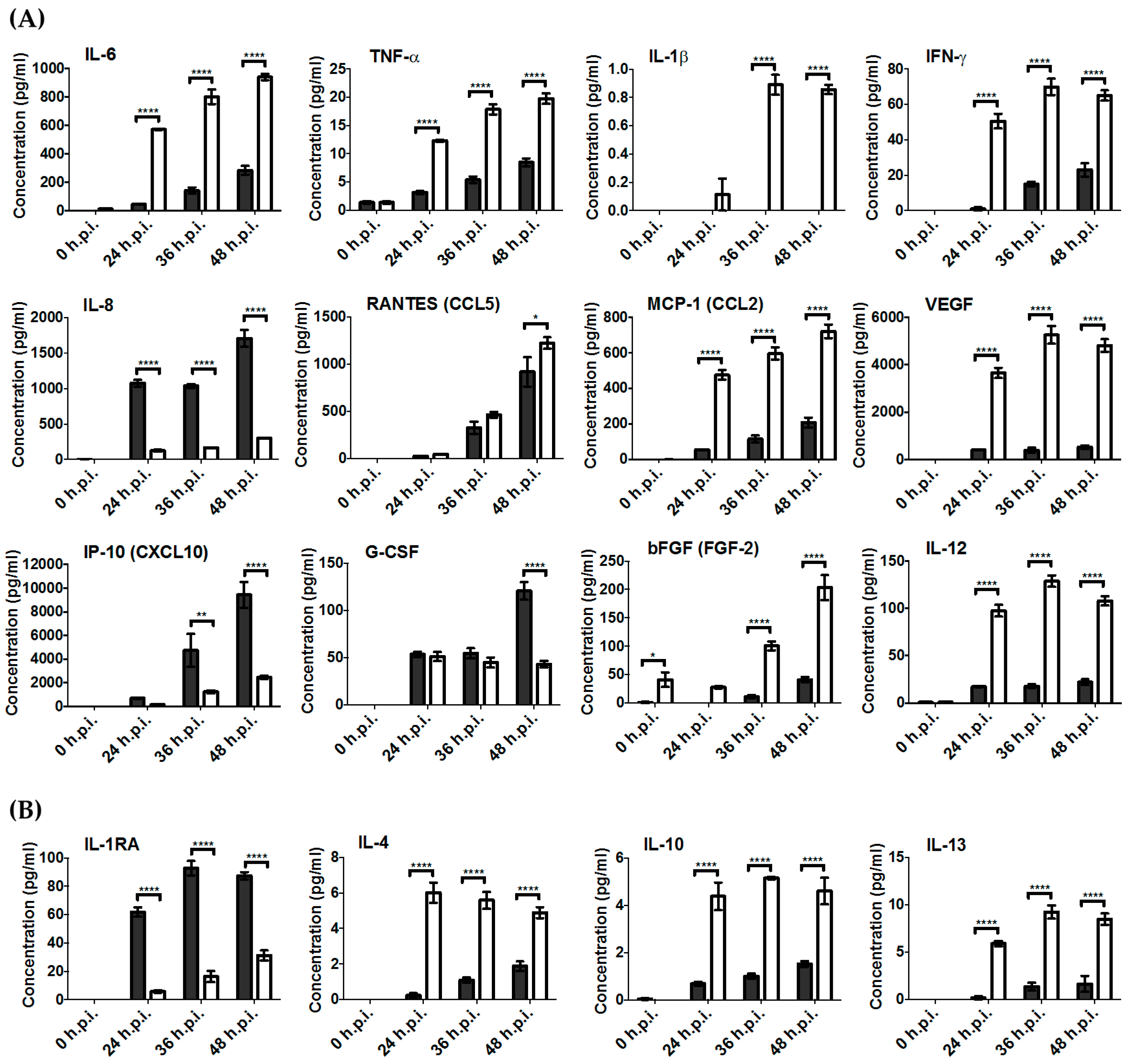
3.4. Key Pro-Inflammatory Soluble Mediators Such as IL-6, MCP-1, RANTES, IFN-γ, and TNF-α Were Produced during RRV Infection
4. Discussion
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Alpert, E.; Isselbacher, K.J.; Schur, P.H. The Pathogenesis of Arthritis Associated with Viral Hepatitis. N. Engl. J. Med. 1971, 285, 185–189. [Google Scholar] [CrossRef] [PubMed]
- Patti, A.M.; Gabriele, A.; Vulcano, A.; Ramieri, M.T.; Rocca, D.C. Effect of hyaluronic acid on human chondrocyte cell lines from articular cartilage. Tissue Cell 2001, 33, 294–300. [Google Scholar] [CrossRef] [PubMed]
- Mabey, T. Cytokines as biochemical markers for knee osteoarthritis. World J. Orthop. 2015, 6, 95. [Google Scholar] [CrossRef] [PubMed]
- Ng, L.F.P.; Chow, A.; Sun, Y.-J.; Kwek, D.J.C.; Lim, P.-L.; Dimatatac, F.; Ng, L.C.; Ooi, E.E.; Choo, K.-H.; Her, Z.; et al. IL-1β, IL-6, and RANTES as Biomarkers of Chikungunya Severity. PLoS ONE 2009, 4, e4261. [Google Scholar] [CrossRef] [PubMed]
- Larrieu, S.; Pouderoux, N.; Pistone, T.; Filleul, L.; Receveur, M.-C.; Sissoko, D.; Ezzedine, K.; Malvy, D. Factors associated with persistence of arthralgia among chikungunya virus-infected travellers: Report of 42 French cases. J. Clin. Virol. 2010, 47, 85–88. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Foo, S.S.; Rulli, N.E.; Taylor, A.; Sheng, K.C.; Herrero, L.J.; Herring, B.L.; Lidbury, B.A.; Li, R.W.; Walsh, N.C.; et al. Arthritogenic alphaviral infection perturbs osteoblast function and triggers pathologic bone loss. Proc. Natl. Acad. Sci. USA 2014, 111, 6040–6045. [Google Scholar] [CrossRef] [PubMed]
- Goupil, B.A.; McNulty, M.A.; Martin, M.J.; McCracken, M.K.; Christofferson, R.C.; Mores, C.N. Novel Lesions of Bones and Joints Associated with Chikungunya Virus Infection in Two Mouse Models of Disease: New Insights into Disease Pathogenesis. PLoS ONE 2016, 11, e0155243. [Google Scholar] [CrossRef] [PubMed]
- Herrero, L.J.; Foo, S.-S.; Sheng, K.-C.; Chen, W.; Forwood, M.R.; Bucala, R.; Mahalingam, S. Pentosan Polysulfate: A Novel Glycosaminoglycan-Like Molecule for Effective Treatment of Alphavirus-Induced Cartilage Destruction and Inflammatory Disease. J. Virol. 2015, 89, 8063–8076. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, R.J.; Niesters, H.G.M.; Hong, Z.; Strauss, J.H. Infectious RNA transcripts from Ross river virus cDNA clones and the construction and characterization of defined chimeras with Sindbis virus. Virology 1991, 182, 430–441. [Google Scholar] [CrossRef]
- Demircan, K.; Hirohata, S.; Nishida, K.; Hatipoglu, O.F.; Oohashi, T.; Yonezawa, T.; Apte, S.S.; Ninomiya, Y. ADAMTS-9 is synergistically induced by interleukin-1β and tumor necrosis factor α in OUMS-27 chondrosarcoma cells and in human chondrocytes. Arthritis Rheum. 2005, 52, 1451–1460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, I.; Jakob, M.; Schäfer, D.; Dick, W.; Spagnoli, G.; Heberer, M. Quantitative analysis of gene expression in human articular cartilage from normal and osteoarthritic joints. Osteoarthr. Cartil. 2001, 9, 112–118. [Google Scholar] [CrossRef] [PubMed]
- Giannini, S.; Buda, R.; Grigolo, B.; Vannini, F.; de Franceschi, L.; Facchini, A. The detached osteochondral fragment as a source of cells for autologous chondrocyte implantation (ACI) in the ankle joint. Osteoarthr. Cartil. 2005, 13, 601–607. [Google Scholar] [CrossRef] [PubMed]
- Simon, M.J.K.; Niehoff, P.; Kimmig, B.; Wiltfang, J.; Açil, Y. Expression profile and synthesis of different collagen types I, II, III, and V of human gingival fibroblasts, osteoblasts, and SaOS-2 cells after bisphosphonate treatment. Clin. Oral Investig. 2009, 14, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Smith, P.N.; Freeman, C.; Yu, D.; Chen, M.; Gatenby, P.A.; Parish, C.R.; Li, R.W. Heparanase in primary human osteoblasts. J. Orthop. Res. 2010, 28, 1315–1322. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.T.; Lin, Y.T.; Chiang, B.L.; Lin, Y.H.; Hou, S.M. High molecular weight hyaluronic acid down-regulates the gene expression of osteoarthritis-associated cytokines and enzymes in fibroblast-like synoviocytes from patients with early osteoarthritis. Osteoarthr. Cartil. 2006, 14, 1237–1247. [Google Scholar] [CrossRef] [PubMed]
- Zhan, Y.; Abi Saab, W.F.; Modi, N.; Stewart, A.M.; Liu, J.; Chadee, D.N. Mixed lineage kinase 3 is required for matrix metalloproteinase expression and invasion in ovarian cancer cells. Exp. Cell Res. 2012, 318, 1641–1648. [Google Scholar] [CrossRef] [PubMed]
- Rosewell, K.L.; Li, F.; Puttabyatappa, M.; Akin, J.W.; Brännström, M.; Curry, T.E., Jr. Ovarian Expression, Localization, and Function of Tissue Inhibitor of Metalloproteinase 3 (TIMP3) During the Periovulatory Period of the Human Menstrual Cycle. Biol. Reprod. 2013, 89. [Google Scholar] [CrossRef] [PubMed]
- Akkiraju, H.; Nohe, A. Role of Chondrocytes in Cartilage Formation, Progression of Osteoarthritis and Cartilage Regeneration. J. Dev. Biol. 2015, 3, 177–192. [Google Scholar] [CrossRef] [PubMed]
- Nie, D.; Ishikawa, Y.; Guo, Y.; Wu, L.N.Y.; Genge, B.R.; Wuthier, R.E.; Sauer, G.R. Inhibition of terminal differentiation and matrix calcification in cultured avian growth plate chondrocytes by Rous sarcoma virus transformation. J. Cell. Biochem. 1998, 69, 453–462. [Google Scholar] [CrossRef]
- Bendiksen, S.; Martinez-Zubiavrra, I.; Tümmler, C.; Knutsen, G.; Elvenes, J.; Olsen, E.; Olsen, R.; Moens, U. Human Endogenous Retrovirus W Activity in Cartilage of Osteoarthritis Patients. BioMed Res. Int. 2014, 2014, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Yokoo, N.; Saito, T.; Uesugi, M.; Kobayashi, N.; Xin, K.Q.; Okuda, K.; Mizukami, H.; Ozawa, K.; Koshino, T. Repair of articular cartilage defect by autologous transplantation of basic fibroblast growth factor gene-transduced chondrocytes with adeno-associated virus vector. Arthritis Rheum. 2005, 52, 164–170. [Google Scholar] [CrossRef] [PubMed]
- Rudd, P.A.; Wilson, J.; Gardner, J.; Larcher, T.; Babarit, C.; Le, T.T.; Anraku, I.; Kumagai, Y.; Loo, Y.M.; Gale, M.; et al. Interferon Response Factors 3 and 7 Protect against Chikungunya Virus Hemorrhagic Fever and Shock. J. Virol. 2012, 86, 9888–9898. [Google Scholar] [CrossRef] [PubMed]
- Morrison, T.E.; Whitmore, A.C.; Shabman, R.S.; Lidbury, B.A.; Mahalingam, S.; Heise, M.T. Characterization of Ross River Virus Tropism and Virus-Induced Inflammation in a Mouse Model of Viral Arthritis and Myositis. J. Virol. 2005, 80, 737–749. [Google Scholar] [CrossRef] [PubMed]
- Soden, M.; Vasudevan, H.; Roberts, B.; Coelen, R.; Hamlin, G.; Vasudevan, S.; La Brooy, J. Detection of viral ribonucleic acid and histologic analysis of inflamed synovium in Ross River virus infection. Arthritis Rheum. 2000, 43, 365–369. [Google Scholar] [CrossRef]
- Morrison, T.E.; Fraser, R.J.; Smith, P.N.; Mahalingam, S.; Heise, M.T. Complement Contributes to Inflammatory Tissue Destruction in a Mouse Model of Ross River Virus-Induced Disease. J. Virol. 2007, 81, 5132–5143. [Google Scholar] [CrossRef] [PubMed]
- Herrero, L.J.; Nelson, M.; Srikiatkhachorn, A.; Gu, R.; Anantapreecha, S.; Fingerle-Rowson, G.; Bucala, R.; Morand, E.; Santos, L.L.; Mahalingam, S. Critical role for macrophage migration inhibitory factor (MIF) in Ross River virus-induced arthritis and myositis. Proc. Natl. Acad. Sci. USA 2011, 108, 12048–12053. [Google Scholar] [CrossRef] [PubMed]
- Lidbury, B.A.; Simeonovic, C.; Maxwell, G.E.; Marshall, I.D.; Hapel, A.J. Macrophage-induced muscle pathology results in morbidity and mortality for Ross River virus-infected mice. J. Infect. Dis. 2000, 181, 27–34. [Google Scholar] [CrossRef] [PubMed]
| Gene | Primer Sequence | Reference |
|---|---|---|
| ADAMTS4 | F: 5′-AGG CAC TGG GCT ACT ACT AT-3′ R: 5′-GGG ATA GTG ACC ACA TTG TT-3′ | [10] |
| ADAMTS5 | F: 5′-TAT GAC AAG TGC GGA GTA TG-3′ R: 5′-TTC AGG GCT AAA TAG GCA GT-3′ | [10] |
| ACAN | F: 5′-TCG AGG ACA GCG AGG CC-3′ R: 5′-TCG AGG GTG TAG CGT GTA GAG A-3′ | [11] |
| COL1A1 | F: 5′-AGG TGC TGA TGG CTC TCC T-3′ R: 5′-GGA CCA CTT TCA CCC TTG T-3′ | [12] |
| COL2A1 | F: 5′-ATG AGG GCG CGG TAG AGA C-3′ R: 5′-CGG CTT CCA CAC ATC CTT AT-3′ | [13] |
| HPSE | F: 5′-TGG ACC TGG ACT TCT TCA CC-3′ R: 5′-TTG ATT CCT TCT TGG GAT CG-3′ | [14] |
| MMP3 | F: 5′-GAC AAA GGA TAC AAC AGG GAC CAA T-3′ R: 5′-TGA GTG AGT GAT AGA GTG GGT ACA T-3′ | [15] |
| MMP9 | F: 5′-GCC ATT CAC GTC GTC CTT AT-3′ R: 5′-TTG ACA GCG ACA AGA AGT GG-3′ | [16] |
| TIMP3 | F: 5′-ACG ATG GCA AGA TGT ACA CAG G-3′ R: 5′-GGA AGT AAC AAA GCA AGG CAG G-3′ | [17] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lim, E.X.Y.; Supramaniam, A.; Lui, H.; Coles, P.; Lee, W.S.; Liu, X.; Rudd, P.A.; Herrero, L.J. Chondrocytes Contribute to Alphaviral Disease Pathogenesis as a Source of Virus Replication and Soluble Factor Production. Viruses 2018, 10, 86. https://doi.org/10.3390/v10020086
Lim EXY, Supramaniam A, Lui H, Coles P, Lee WS, Liu X, Rudd PA, Herrero LJ. Chondrocytes Contribute to Alphaviral Disease Pathogenesis as a Source of Virus Replication and Soluble Factor Production. Viruses. 2018; 10(2):86. https://doi.org/10.3390/v10020086
Chicago/Turabian StyleLim, Elisa X. Y., Aroon Supramaniam, Hayman Lui, Peta Coles, Wai Suet Lee, Xiang Liu, Penny A. Rudd, and Lara J. Herrero. 2018. "Chondrocytes Contribute to Alphaviral Disease Pathogenesis as a Source of Virus Replication and Soluble Factor Production" Viruses 10, no. 2: 86. https://doi.org/10.3390/v10020086





