Deep Sequencing-Based Transcriptome Profiling Reveals Avian Interferon-Stimulated Genes and Provides Comprehensive Insight into Newcastle Disease Virus-Induced Host Responses
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell lines and Viral Propagation
2.2. Viral Infection
2.3. Total RNA Extraction
2.4. RNA-Sequencing
2.5. Transcriptome Assembly
2.6. Coding Potential Analysis
2.7. Expression Analysis
2.8. GO Enrichment and KEGG Pathway Analysis
2.9. ISGs Induced by NDV Infection
2.10. Validation of RNA-Seq Data by Quantitative Real-Time PCR (qPCR) Analysis
3. Results
3.1. Identification of Coding Transcripts in NDV-Infected CEFs
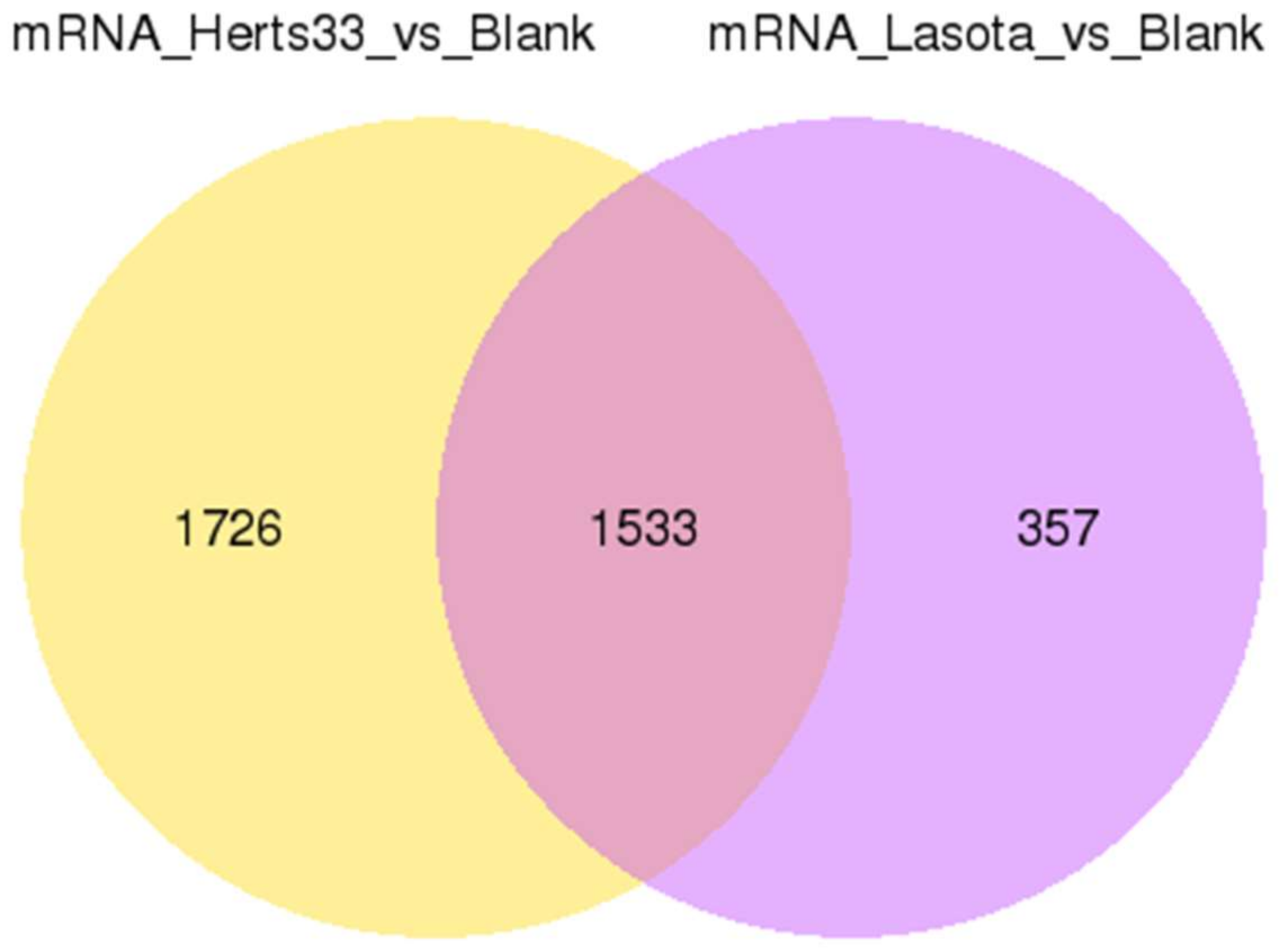
3.2. Global Changes in Expression in Response to NDV Infection
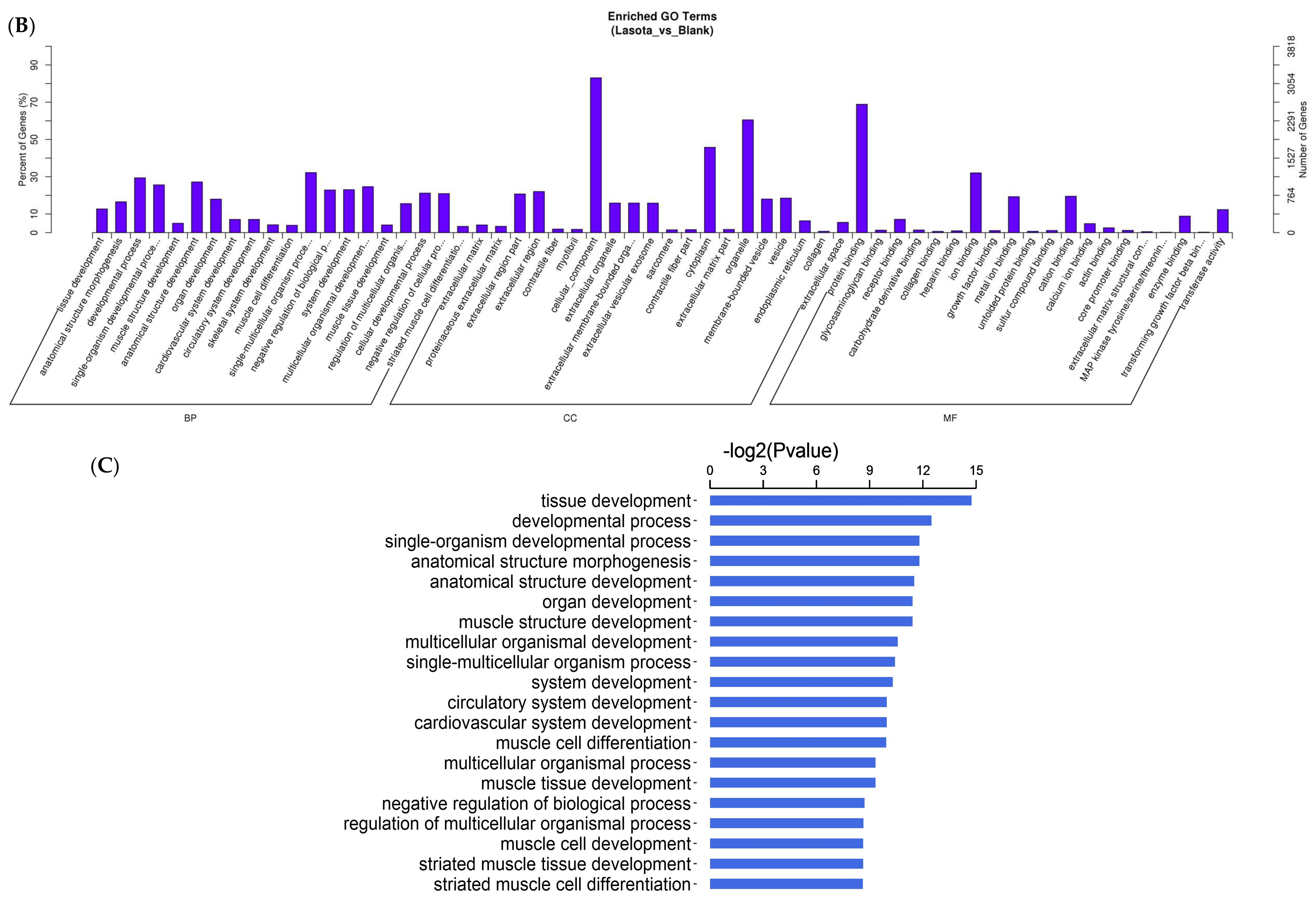
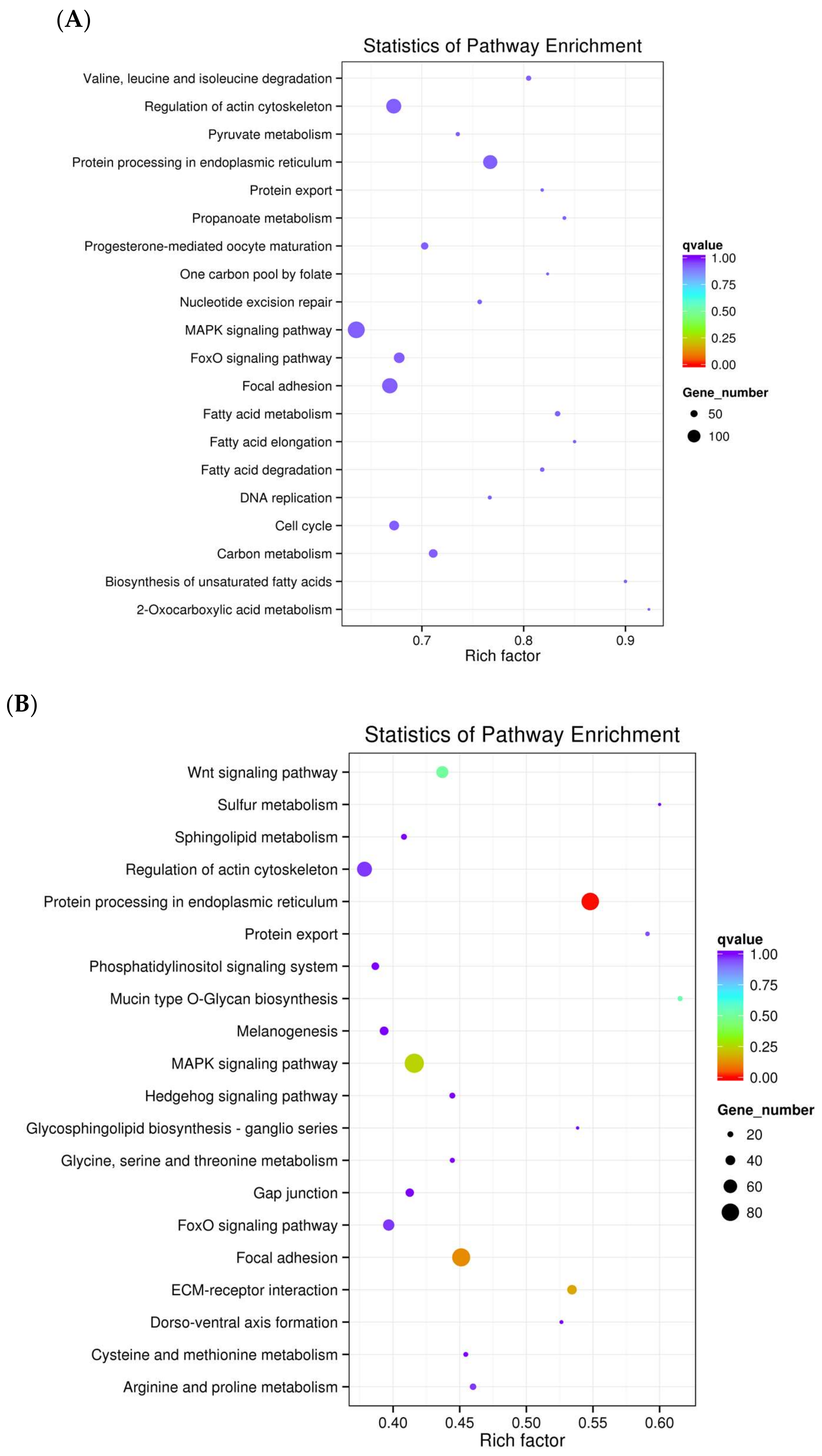
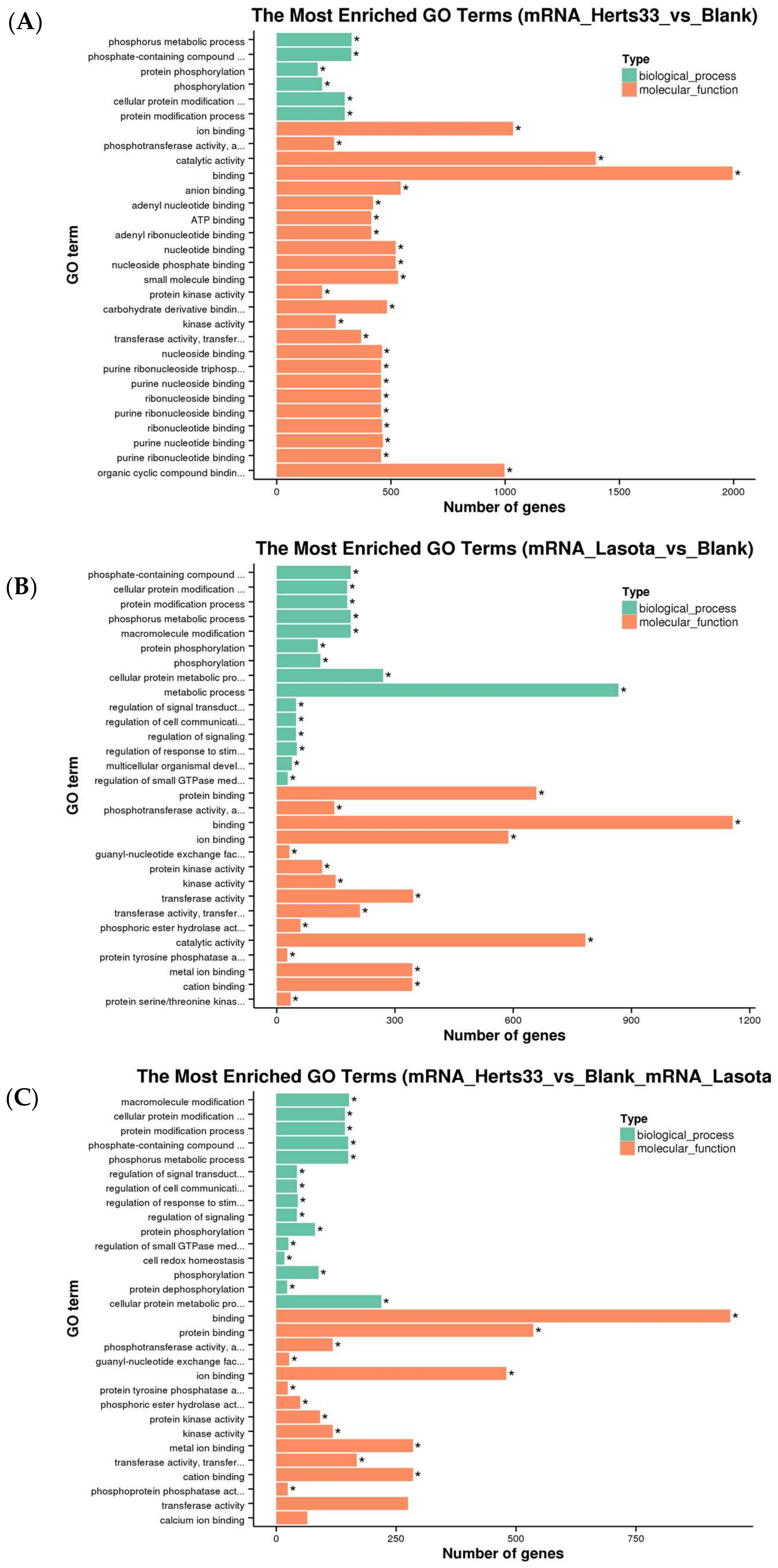
3.3. Functional Analysis of Differentially Expressed Transcripts
3.4. Analysis of Chicken ISGs in NDV-Infected CEFs
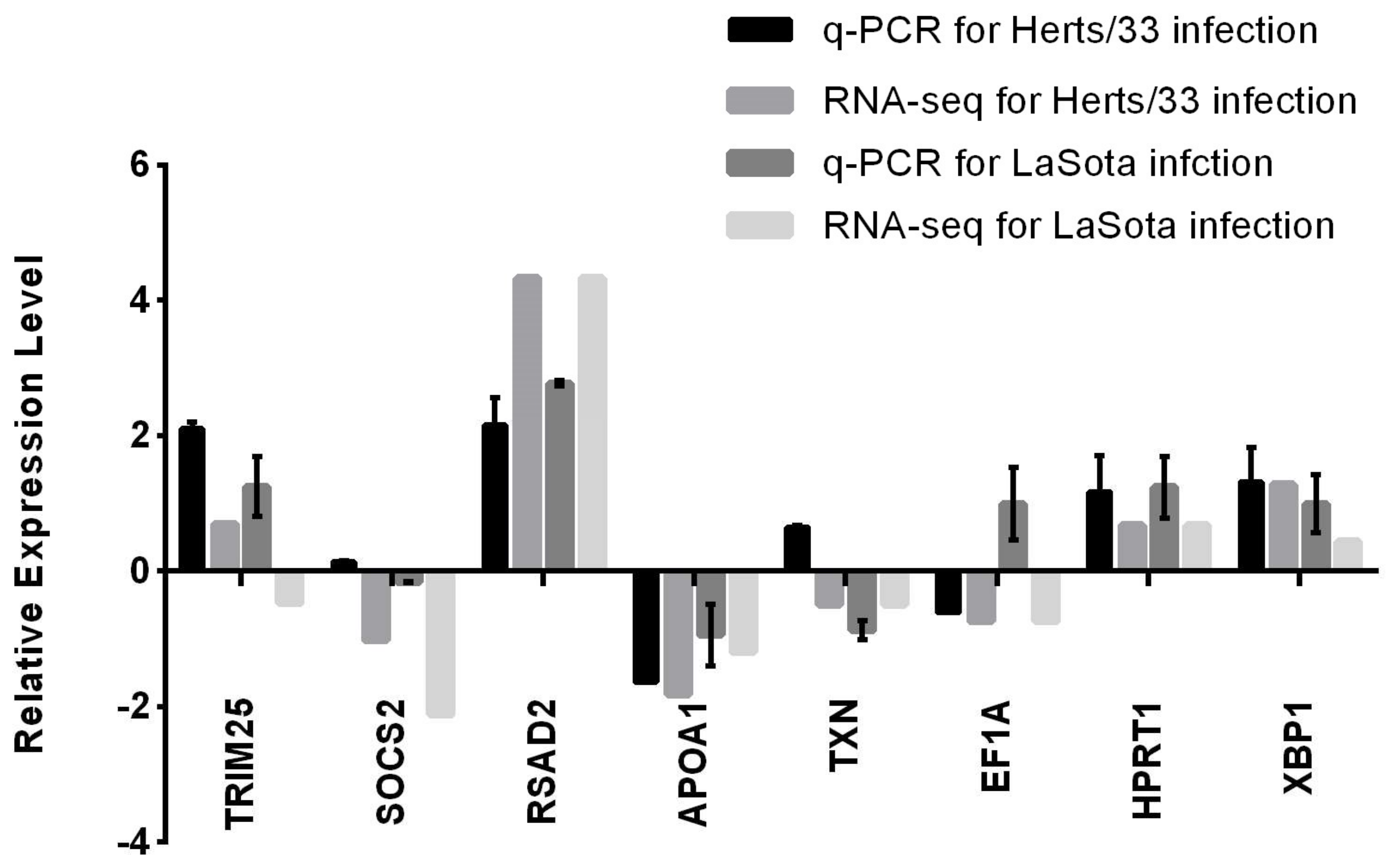
3.5. Validation of RNA-Seq Data by Quantitative Real-Time PCR (qPCR)
4. Discussion
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Lancaster, J. A history of Newcastle disease with comments on its economic effects. World Poult. Sci. J. 1976, 32, 167–175. [Google Scholar] [CrossRef]
- Alexander, S.D. Newcastle disease, other Avian paramyxoviruses, and pneumovirus infections. Dis. Poult. 2008, 357, 75–98. [Google Scholar]
- Dortmans, J.C.; Koch, G.; Rottier, P.J.; Peeters, B.P. Virulence of Newcastle disease virus: What is known so far? Vet. Res. 2011, 42, 122. [Google Scholar] [CrossRef] [PubMed]
- Spradbrow, P. Geographical distribution. In Newcastle Disease; Alexander, D.J., Ed.; Kluwer Academic Publishers: Boston, MA, USA, 1988; pp. 247–255. [Google Scholar]
- World Organisation for Animal Health. Newcastle disease. In OIE Terrestrial Manual 2012: Manual of Diagnostic Tests and Vaccines for Terrestrial Animals; World Organisation for Animal Health: Paris, France, 2012; Chapter 2.3.14; pp. 576–589. [Google Scholar]
- Chambers, P.; Millar, N.S.; Bingham, R.W.; Emmerson, P.T. Molecular cloning of complementary DNA to Newcastle disease virus, and nucleotide sequence analysis of the junction between the genes encoding the haemagglutinin-neuraminidase and the large protein. J. Gen. Virol. 1986, 67, 475–486. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, L.A.; Bowie, A.G. Sensing and signaling in antiviral innate immunity. Curr. Biol. 2010, 20, R328–R333. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, O.; Akira, S. Pattern recognition receptors and inflammation. Cell 2010, 140, 805–820. [Google Scholar] [CrossRef] [PubMed]
- Randall, R.E.; Goodbourn, S. Interferons and viruses: An interplay between induction, signalling, antiviral responses and virus countermeasures. J. Gen. Virol. 2008, 89, 1–47. [Google Scholar] [CrossRef] [PubMed]
- Schneider, W.M.; Chevillotte, M.D.; Rice, C.M. Interferon-stimulated genes: A complex web of host defenses. Annu. Rev. Immunol. 2014, 32, 513–545. [Google Scholar] [CrossRef] [PubMed]
- Isaacs, A.; Lindenmann, J. Pillars article: Virus interference. I. The interferon. Proc. R. Soc. Lond. B Biol. Sci. 1957, 147, 258–267. [Google Scholar] [CrossRef] [PubMed]
- Sekellick, M.J.; Ferrandino, A.F.; Hopkins, D.A.; Marcus, P.I. Chicken interferon gene: Cloning, expression, and analysis. J. Interferon Res. 1994, 14, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.; Feng, M.; Zhao, X.; Dai, X.; Xiang, B.; Gao, P.; Li, Y.; Li, Y.; Ren, T. Newcastle disease virus infection in chicken embryonic fibroblasts but not duck embryonic fibroblasts is associated with elevated host innate immune response. Virol. J. 2016, 13, 41. [Google Scholar] [CrossRef] [PubMed]
- Kristeen-Teo, Y.W.; Yeap, S.K.; Tan, S.W.; Omar, A.R.; Ideris, A.; Tan, S.G.; Alitheen, N.B. The effects of different velogenic NDV infections on the chicken bursa of fabricius. BMC Vet. Res. 2017, 13, 151. [Google Scholar] [CrossRef] [PubMed]
- Susta, L.; Cornax, I.; Diel, D.G.; Garcia, S.C.; Miller, P.J.; Liu, X.; Hu, S.; Brown, C.C.; Afonso, C.L. Expression of interferon γ by a highly virulent strain of Newcastle disease virus decreases its pathogenicity in chickens. Microb. Pathog. 2013, 61–62, 73–83. [Google Scholar] [CrossRef] [PubMed]
- Xiang, B.; Zhu, W.; Li, Y.; Gao, P.; Liang, J.; Liu, D.; Ding, C.; Liao, M.; Kang, Y.; Ren, T. Immune responses of mature chicken bone-marrow-derived dendritic cells infected with Newcastle disease virus strains with differing pathogenicity. Arch. Virol. 2018, 2018, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Giotis, E.S.; Robey, R.C.; Skinner, N.G.; Tomlinson, C.D.; Goodbourn, S.; Skinner, M.A. Chicken interferome: Avian interferon-stimulated genes identified by microarray and RNA-seq of primary chick embryo fibroblasts treated with a chicken type I interferon (IFN-α). Vet. Res. 2016, 47, 75. [Google Scholar] [CrossRef] [PubMed]
- Ravindra, P.V.; Tiwari, A.K.; Ratta, B.; Chaturvedi, U.; Palia, S.K.; Chauhan, R.S. Newcastle disease virus-induced cytopathic effect in infected cells is caused by Apoptosis. Virus Res. 2009, 141, 13–20. [Google Scholar] [CrossRef] [PubMed]
- Lamb, R.A.; Kolakofsky, D. Paramyxoviridae: The viruses and their replication. In Fields Virology, 3rd ed.; Fields, B.N., Knipe, D.M., Howley, P.M., Eds.; Lippincott Raven: Philadelphia, PA, USA, 1996; Volume 1, pp. 1177–1203. [Google Scholar]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-SEQ reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [PubMed]
- Guttman, M.; Garber, M.; Levin, J.Z.; Donaghey, J.; Robinson, J.; Adiconis, X.; Fan, L.; Koziol, M.J.; Gnirke, A.; Nusbaum, C.; et al. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-Exonic structure of lincRNAs. Nat. Biotechnol. 2010, 28, 503–510. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSEQ—A python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Luo, H.; Bu, D.; Zhao, G.; Yu, K.; Zhang, C.; Liu, Y.; Chen, R.; Zhao, Y. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res. 2013, 41, e166. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.F.; Jungreis, I.; Kellis, M. PhyloCSF: A comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics 2011, 27, i275–i282. [Google Scholar] [CrossRef] [PubMed]
- Bateman, A.; Birney, E.; Cerruti, L.; Durbin, R.; Etwiller, L.; Eddy, S.R.; Griffiths-Jones, S.; Howe, K.L.; Marshall, M.; Sonnhammer, E.L. The PFAM protein families database. Nucleic Acids Res. 2002, 30, 276–280. [Google Scholar] [CrossRef] [PubMed]
- Punta, M.; Coggill, P.C.; Eberhardt, R.Y.; Mistry, J.; Tate, J.; Boursnell, C.; Pang, N.; Forslund, K.; Ceric, G.; Clements, J.; et al. The PFAM protein families database. Nucleic Acids Res. 2012, 40, D290–D301. [Google Scholar] [CrossRef] [PubMed]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-SEQ: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.; Cai, T.; Olyarchuk, J.G.; Wei, L. Automated genome annotation and pathway identification using the kegg orthology (KO) as a controlled vocabulary. Bioinformatics 2005, 21, 3787–3793. [Google Scholar] [CrossRef] [PubMed]
- Rusinova, I.; Forster, S.; Yu, S.; Kannan, A.; Masse, M.; Cumming, H.; Chapman, R.; Hertzog, P.J. Interferome v2.0: An updated database of annotated interferon-regulated genes. Nucleic Acids Res. 2013, 41, D1040–D1046. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Hua, S.; Chen, H.R.; Ouyang, Z.; Einkauf, K.; Tse, S.; Ard, K.; Ciaranello, A.; Yawetz, S.; Sax, P.; et al. Transcriptional changes during naturally acquired zika virus infection render dendritic cells highly conducive to viral replication. Cell Rep. 2017, 21, 3471–3482. [Google Scholar] [CrossRef] [PubMed]
- Zhong, H.; Hu, X.; Janowski, A.B.; Storch, G.A.; Su, L.; Cao, L.; Yu, J. Whole transcriptome profiling reveals major cell types in the cellular immune response against acute and chronic active Epstein-Barr virus infection. Sci. Rep. 2017, 7, 17775. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.; Caviness, K.; Buehler, J.; Smithey, M.; Nikolich-Zugich, J.; Goodrum, F. Transcriptome-wide characterization of human cytomegalovirus in natural infection and experimental latency. Proc. Natl. Acad. Sci. USA 2017, 114, E10586–E10595. [Google Scholar] [CrossRef] [PubMed]
- Deist, M.S.; Gallardo, R.A.; Bunn, D.A.; Kelly, T.R.; Dekkers, J.C.M.; Zhou, H.; Lamont, S.J. Novel mechanisms revealed in the trachea transcriptome of resistant and susceptible chicken lines following infection with Newcastle disease virus. Clin. Vaccine Immunol. 2017, 24, e00027-17. [Google Scholar] [CrossRef] [PubMed]
- Deist, M.S.; Gallardo, R.A.; Bunn, D.A.; Dekkers, J.C.M.; Zhou, H.; Lamont, S.J. Resistant and susceptible chicken lines show distinctive responses to Newcastle disease virus infection in the lung transcriptome. BMC Genom. 2017, 18, 989. [Google Scholar] [CrossRef] [PubMed]
- Goodwin, C.M.; Xu, S.; Munger, J. Stealing the keys to the kitchen: Viral manipulation of the host cell metabolic network. Trends Microbiol. 2015, 23, 789–798. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, E.L.; Lagunoff, M. Viral activation of cellular metabolism. Virology 2015, 479–480, 609–618. [Google Scholar] [CrossRef] [PubMed]
- Fontaine, K.A.; Sanchez, E.L.; Camarda, R.; Lagunoff, M. Dengue virus induces and requires glycolysis for optimal replication. J. Virol. 2015, 89, 2358–2366. [Google Scholar] [CrossRef] [PubMed]
- Chambers, J.W.; Maguire, T.G.; Alwine, J.C. Glutamine metabolism is essential for human cytomegalovirus infection. J. Virol. 2010, 84, 1867–1873. [Google Scholar] [CrossRef] [PubMed]
- Thai, M.; Graham, N.A.; Braas, D.; Nehil, M.; Komisopoulou, E.; Kurdistani, S.K.; McCormick, F.; Graeber, T.G.; Christofk, H.R. Adenovirus E4ORF1-induced MYC activation promotes host cell anabolic glucose metabolism and virus replication. Cell Metab. 2014, 19, 694–701. [Google Scholar] [CrossRef] [PubMed]
- Hand, R. Thymidine metabolism and DNA synthesis in Newcastle disease virus-infected cells. J. Virol. 1976, 19, 801–809. [Google Scholar] [PubMed]
- Sijtsma, S.R.; West, C.E.; Rombout, J.H.; van der Zijpp, A.J. Effect of Newcastle disease virus infection on Vitamin A metabolism in chickens. J. Nutr. 1989, 119, 940–947. [Google Scholar] [CrossRef] [PubMed]
- Venkata Subbaiah, K.C.; Valluru, L.; Rajendra, W.; Ramamurthy, C.; Thirunavukkarusu, C.; Subramanyam, R. Newcastle disease virus (NDV) induces protein oxidation and nitration in brain and liver of chicken: Ameliorative effect of Vitamin E. Int. J. Biochem. Cell Biol. 2015, 64, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Pearson, G.; Robinson, F.; Beers Gibson, T.; Xu, B.E.; Karandikar, M.; Berman, K.; Cobb, M.H. Mitogen-activated protein (MAP) kinase pathways: Regulation and physiological functions. Endocr. Rev. 2001, 22, 153–183. [Google Scholar] [CrossRef] [PubMed]
- Pleschka, S. RNA viruses and the mitogenic RAF/MEK/ERK signal transduction cascade. Biol. Chem. 2008, 389, 1273–1282. [Google Scholar] [CrossRef] [PubMed]
- Ch’ng, W.C.; Abd-Aziz, N.; Ong, M.H.; Stanbridge, E.J.; Shafee, N. Human renal carcinoma cells respond to Newcastle disease virus infection through activation of the p38 MAPK/NF-κβ/Iκβα pathway. Cell. Oncol. 2015, 38, 279–288. [Google Scholar] [CrossRef] [PubMed]
- Meng, S.; Zhou, Z.; Chen, F.; Kong, X.; Liu, H.; Jiang, K.; Liu, W.; Hu, M.; Zhang, X.; Ding, C.; et al. Newcastle disease virus induces apoptosis in Cisplatin-resistant human lung adenocarcinoma A549 cells in vitro and in vivo. Cancer Lett. 2012, 317, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Bian, J.; Wang, K.; Kong, X.; Liu, H.; Chen, F.; Hu, M.; Zhang, X.; Jiao, X.; Ge, B.; Wu, Y.; et al. Caspase- and p38-MAPK-dependent induction of apoptosis in A549 lung cancer cells by Newcastle disease virus. Arch. Virol. 2011, 156, 1335–1344. [Google Scholar] [CrossRef] [PubMed]
- Reid, D.W.; Campos, R.K.; Child, J.R.; Zheng, T.; Chan, K.W.K.; Bradrick, S.S.; Vasudevan, S.G.; Garcia-Blanco, M.A.; Nicchitta, C.V. Dengue virus selectively annexes endoplasmic reticulum-associated translation machinery as a strategy for co-opting host cell protein synthesis. J. Virol. 2018, 92, e01766-17. [Google Scholar] [CrossRef] [PubMed]
- Boson, B.; Granio, O.; Bartenschlager, R.; Cosset, F.L. A concerted action of Hepatitis C virus P7 and nonstructural protein 2 regulates core localization at the endoplasmic reticulum and virus assembly. PLoS Pathog. 2011, 7, e1002144. [Google Scholar] [CrossRef] [PubMed]
- Inoue, T.; Tsai, B. How viruses use the endoplasmic reticulum for entry, replication, and assembly. Cold Spring Harb. Perspect. Biol. 2013, 5, a013250. [Google Scholar] [CrossRef] [PubMed]
- Bu, X.; Zhao, Y.; Zhang, Z.; Wang, M.; Li, M.; Yan, Y. Recombinant Newcastle disease virus (RL-RVG) triggers autophagy and apoptosis in gastric carcinoma cells by inducing ER stress. Am. J. Cancer Res. 2016, 6, 924–936. [Google Scholar] [PubMed]
- Cheng, J.H.; Sun, Y.J.; Zhang, F.Q.; Zhang, X.R.; Qiu, X.S.; Yu, L.P.; Wu, Y.T.; Ding, C. Newcastle disease virus NP and P proteins induce autophagy via the endoplasmic reticulum stress-related unfolded protein response. Sci. Rep. 2016, 6, 24721. [Google Scholar] [CrossRef] [PubMed]
- Fabian, Z.; Csatary, C.M.; Szeberenyi, J.; Csatary, L.K. P53-independent endoplasmic reticulum stress-mediated cytotoxicity of a Newcastle disease virus strain in tumor cell lines. J. Virol. 2007, 81, 2817–2830. [Google Scholar] [CrossRef] [PubMed]
- Rue, C.A.; Susta, L.; Cornax, I.; Brown, C.C.; Kapczynski, D.R.; Suarez, D.L.; King, D.J.; Miller, P.J.; Afonso, C.L. Virulent newcastle disease virus elicits a strong innate immune response in chickens. J. Gen. Virol. 2011, 92, 931–939. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.Q.; Tian, M.X.; Wang, Y.P.; Zhao, Y.; Zou, N.L.; Zhao, F.F.; Cao, S.J.; Wen, X.T.; Liu, P.; Huang, Y. The different expression of immune-related cytokine genes in response to velogenic and lentogenic newcastle disease viruses infection in chicken peripheral blood. Mol. Biol. Rep. 2012, 39, 3611–3618. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Hu, J.; Hu, S.; Liu, X.; Wang, X.; Zhu, J.; Liu, X. Strong innate immune response and cell death in chicken splenocytes infected with genotype viid newcastle disease virus. Virol. J. 2012, 9, 208. [Google Scholar] [CrossRef] [PubMed]


| Gene Name | Forward Primer (5′–3′) | Reverse Primer (5′–3′) |
|---|---|---|
| TRIM25 | TCAAGAGTCCCACCCTTCCA | AGCAGCTCAATGGACAGCAT |
| SOSC2 | GCGCGCAGGGTGGTACT | ATGCGAACTGTCCCTAACCAA |
| RSAD2 | ACACCTCAGGGAATCACCCTTT | AAGGATTCTCTGTTATCCAAGCTGAA |
| APOA1 | GATGCCATCGCCCAGTTC | CCATGTCCTCACGCAGCTT |
| TXN | GTCTGTGTGACAAGTTTGGTGATG | AATGTTGGCATGCACTTCACAT |
| EF1A | GGGCACCTCATCTACAAATGC | ACCCAGGCGTATTTGAAGGA |
| XBP1 | GTGCGAGTCTACGGATGTGAAG | CTGCAGAGGAACACGTAGTCTGA |
| HPRT1 | CCAAACATTATGCAGACGATCTG | CCCATGCCCTTCATAATTTCA |
| GAPDH | CAATGATCCCTTCATCGATCTG | TTTCCCGTTCTCAGCCTTGA |
| Category | Control_1 | Control_2 | Control_3 | Herts/33_1 | Herts/33_2 | Herts/33_3 | LaSota_1 | LaSota_2 | LaSota_3 |
|---|---|---|---|---|---|---|---|---|---|
| Raw reads | 109,126,046 | 110,981,726 | 86,062,988 | 83,496,140 | 94,400,454 | 93,150,904 | 93,479,188 | 83,743,856 | 87,401,712 |
| Clean reads | 104,687,082 | 106,667,980 | 81,878,214 | 80,208,914 | 90,581,186 | 89,441,660 | 89,772,806 | 80,667,246 | 84,119,752 |
| Clean bases | 15.7 G | 16 G | 12.28 G | 12.03 G | 13.59 G | 13.42 G | 13.47 G | 12.1 G | 12.62 G |
| Total mapped | 89,050,776 | 91,232,554 | 68,861,314 | 52,421,734 | 59,372,721 | 61,229,723 | 72,781,410 | 65,496,022 | 68,125,651 |
| 85.06% | 85.53% | 84.1% | 65.36% | 65.55% | 68.46% | 81.07% | 81.19% | 80.99% | |
| Protein coding | 32,050,717 | 32,705,773 | 25,620,258 | 18,519,665 | 21,280,793 | 21,712,088 | 26,453,027 | 24,048,464 | 24,471,856 |
| 75.36% | 75.45% | 75.57% | 74.08% | 73.94% | 73.54% | 75.13% | 75.16% | 74.78% |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, W.; Qiu, X.; Song, C.; Sun, Y.; Meng, C.; Liao, Y.; Tan, L.; Ding, Z.; Liu, X.; Ding, C. Deep Sequencing-Based Transcriptome Profiling Reveals Avian Interferon-Stimulated Genes and Provides Comprehensive Insight into Newcastle Disease Virus-Induced Host Responses. Viruses 2018, 10, 162. https://doi.org/10.3390/v10040162
Liu W, Qiu X, Song C, Sun Y, Meng C, Liao Y, Tan L, Ding Z, Liu X, Ding C. Deep Sequencing-Based Transcriptome Profiling Reveals Avian Interferon-Stimulated Genes and Provides Comprehensive Insight into Newcastle Disease Virus-Induced Host Responses. Viruses. 2018; 10(4):162. https://doi.org/10.3390/v10040162
Chicago/Turabian StyleLiu, Weiwei, Xusheng Qiu, Cuiping Song, Yingjie Sun, Chunchun Meng, Ying Liao, Lei Tan, Zhuang Ding, Xiufan Liu, and Chan Ding. 2018. "Deep Sequencing-Based Transcriptome Profiling Reveals Avian Interferon-Stimulated Genes and Provides Comprehensive Insight into Newcastle Disease Virus-Induced Host Responses" Viruses 10, no. 4: 162. https://doi.org/10.3390/v10040162
APA StyleLiu, W., Qiu, X., Song, C., Sun, Y., Meng, C., Liao, Y., Tan, L., Ding, Z., Liu, X., & Ding, C. (2018). Deep Sequencing-Based Transcriptome Profiling Reveals Avian Interferon-Stimulated Genes and Provides Comprehensive Insight into Newcastle Disease Virus-Induced Host Responses. Viruses, 10(4), 162. https://doi.org/10.3390/v10040162




