Neutralizing Monoclonal Anti-SARS-CoV-2 Antibodies Isolated from Immunized Rabbits Define Novel Vulnerable Spike-Protein Epitope
Abstract
:1. Introduction
2. Materials and Methods
2.1. Expression of SARS-CoV-2 Spike Recombinant Protein
2.2. Rabbits Immunization
2.3. ELISA
2.4. Isolation of Peripheral Blood Mononuclear Cells and Splenocytes
2.5. Isolation and Ig Gene Amplification of Single B Cells
2.6. Production of scFv-Fc Antibodies
2.7. Cells
2.8. Plaque Reduction Neutralization Test (PRNT)
2.9. Biolayer Interferometry
2.10. Epitope Mapping
3. Results
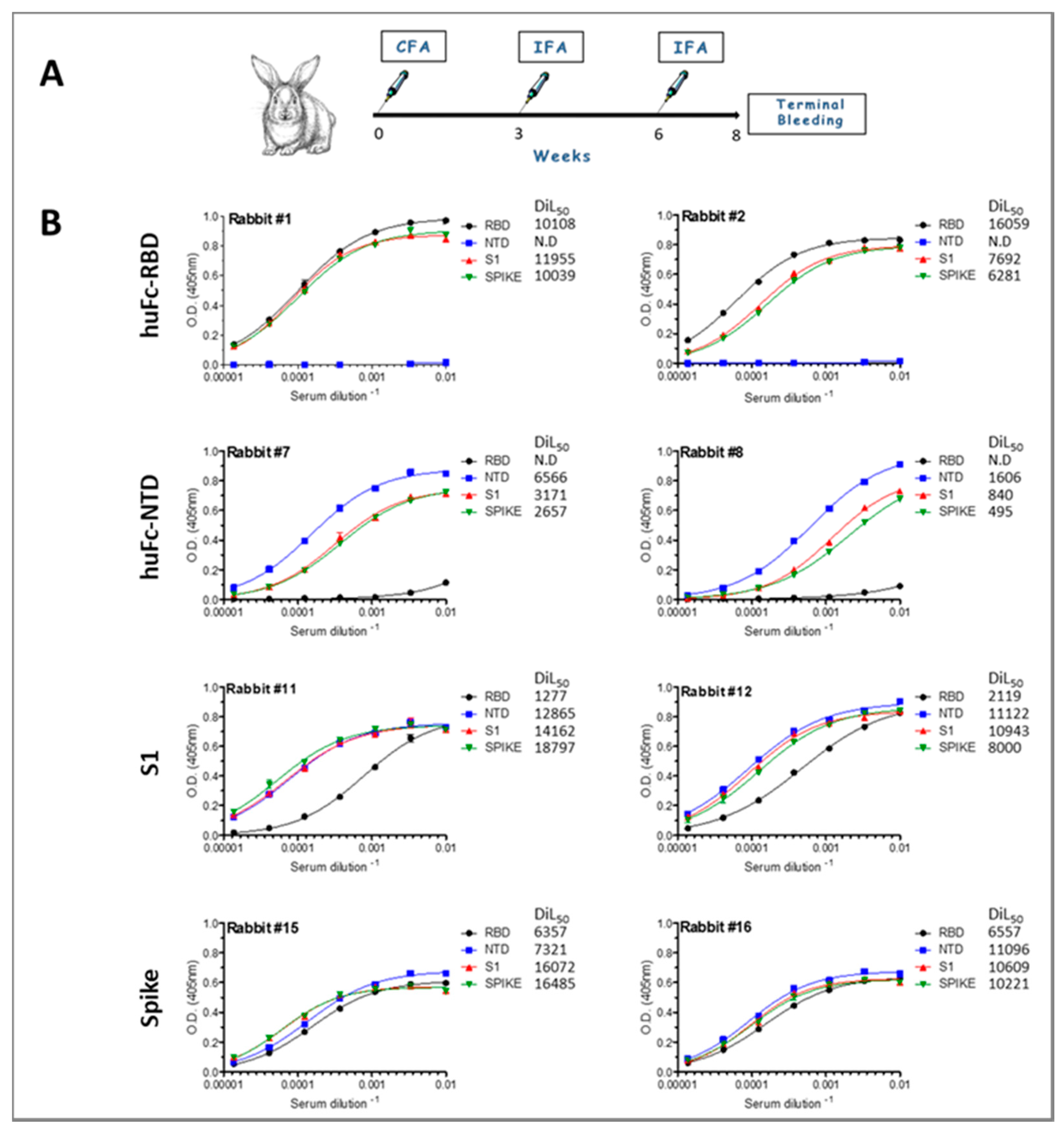
3.1. Immunization of Rabbits and Characterization of the Humoral Immune Response
3.2. Isolation of Anti-RBD Antibodies
3.3. Isolation of Anti-NTD Antibodies
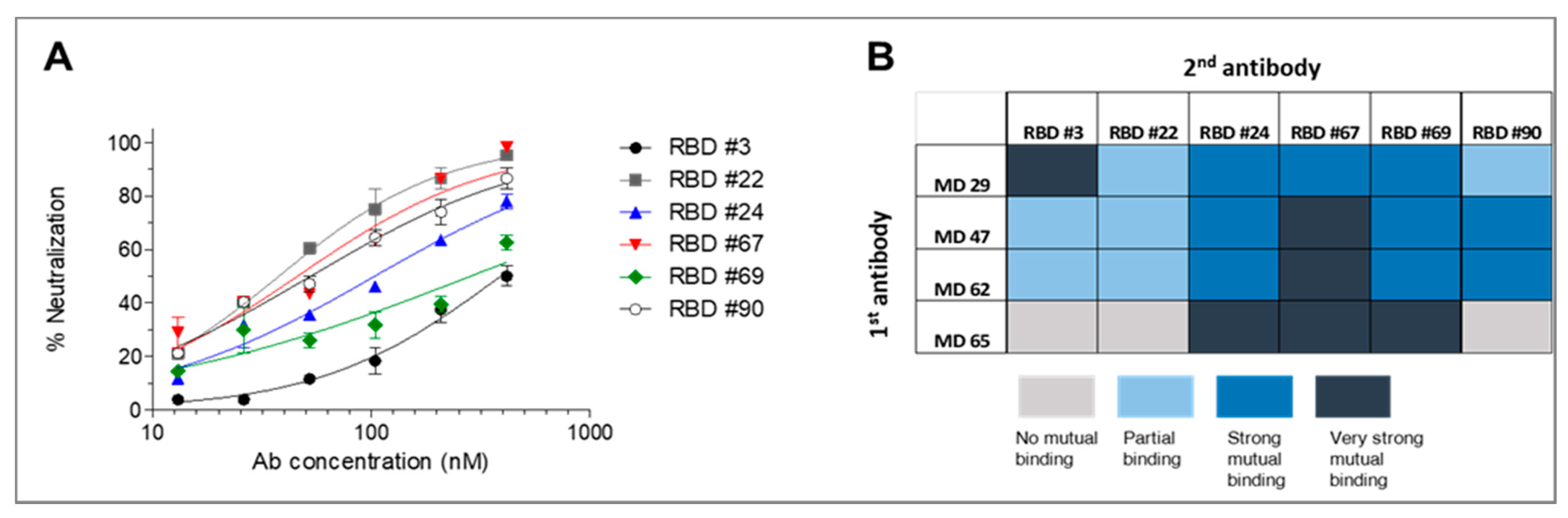
3.4. Neutralizing Potency of the Selected mAbs
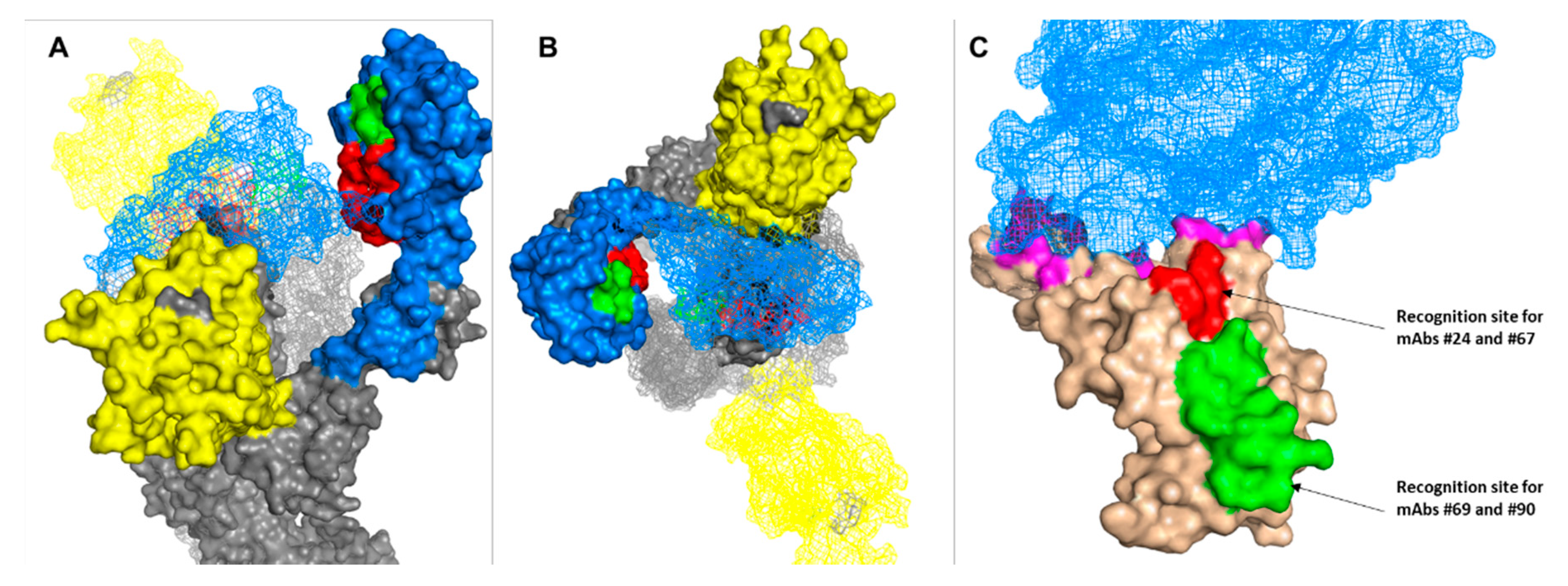
3.5. Epitope Mapping
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hoffmann, M.; Kleine-Weber, H.; Pohlmann, S. A Multibasic Cleavage Site in the Spike Protein of SARS-CoV-2 Is Essential for Infection of Human Lung Cells. Mol. Cell 2020, 78, 779–784.e5. [Google Scholar] [CrossRef] [PubMed]
- Walls, A.C.; Park, Y.J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 183, 1735. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Kruger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef]
- Alsoussi, W.B.; Turner, J.S.; Case, J.B.; Zhao, H.; Schmitz, A.J.; Zhou, J.Q.; Chen, R.E.; Lei, T.; Rizk, A.A.; McIntire, K.M.; et al. A Potently Neutralizing Antibody Protects Mice against SARS-CoV-2 Infection. J. Immunol. 2020, 205, 915–922. [Google Scholar] [CrossRef]
- Bonam, S.R.; Kotla, N.G.; Bohara, R.A.; Rochev, Y.; Webster, T.J.; Bayry, J. Potential immuno-nanomedicine strategies to fight COVID-19 like pulmonary infections. Nano Today 2021, 36, 101051. [Google Scholar] [CrossRef] [PubMed]
- Case, J.B.; Rothlauf, P.W.; Chen, R.E.; Liu, Z.; Zhao, H.; Kim, A.S.; Bloyet, L.M.; Zeng, Q.; Tahan, S.; Droit, L.; et al. Neutralizing Antibody and Soluble ACE2 Inhibition of a Replication-Competent VSV-SARS-CoV-2 and a Clinical Isolate of SARS-CoV-2. SSRN 2020, 3606354. [Google Scholar] [CrossRef]
- Hassan, A.O.; Case, J.B.; Winkler, E.S.; Thackray, L.B.; Kafai, N.M.; Bailey, A.L.; McCune, B.T.; Fox, J.M.; Chen, R.E.; Alsoussi, W.B.; et al. A SARS-CoV-2 Infection Model in Mice Demonstrates Protection by Neutralizing Antibodies. Cell 2020, 182, 744–753.e4. [Google Scholar] [CrossRef] [PubMed]
- Noy-Porat, T.; Mechaly, A.; Levy, Y.; Makdasi, E.; Alcalay, R.; Gur, D.; Aftalion, M.; Falach, R.; Leviatan Ben-Arye, S.; Lazar, S.; et al. Therapeutic antibodies, targeting the SARS-CoV-2 spike N-terminal domain, protect lethally infected K18-hACE2 mice. bioRxiv 2021, 428995. [Google Scholar] [CrossRef]
- Rogers, T.F.; Zhao, F.; Huang, D.; Beutler, N.; Burns, A.; He, W.T.; Limbo, O.; Smith, C.; Song, G.; Woehl, J.; et al. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science 2020, 369, 956–963. [Google Scholar] [CrossRef] [PubMed]
- Rosenfeld, R.; Noy-Porat, T.; Mechaly, A.; Makdasi, E.; Levy, Y.; Alcalay, R.; Falach, R.; Aftalion, M.; Epstein, E.; Gur, D.; et al. Post-exposure protection of SARS-CoV-2 lethal infected K18-hACE2 transgenic mice by neutralizing human monoclonal antibody. Nat. Commun. 2021, 12, 944. [Google Scholar] [CrossRef]
- Zost, S.J.; Gilchuk, P.; Case, J.B.; Binshtein, E.; Chen, R.E.; Nkolola, J.P.; Schafer, A.; Reidy, J.X.; Trivette, A.; Nargi, R.S.; et al. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature 2020, 584, 443–449. [Google Scholar] [CrossRef] [PubMed]
- Abbasi, J. COVID-19 Antibody Trials Have Begun. JAMA 2020, 324, 128. [Google Scholar] [CrossRef] [PubMed]
- Mahase, E. Covid-19: RECOVERY trial will evaluate “antiviral antibody cocktail”. BMJ 2020, 370, m3584. [Google Scholar] [CrossRef]
- Tuccori, M.; Ferraro, S.; Convertino, I.; Cappello, E.; Valdiserra, G.; Blandizzi, C.; Maggi, F.; Focosi, D. Anti-SARS-CoV-2 neutralizing monoclonal antibodies: Clinical pipeline. MAbs 2020, 12, 1854149. [Google Scholar] [CrossRef] [PubMed]
- Brouwer, P.J.M.; Caniels, T.G.; van der Straten, K.; Snitselaar, J.L.; Aldon, Y.; Bangaru, S.; Torres, J.L.; Okba, N.M.A.; Claireaux, M.; Kerster, G.; et al. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science 2020, 369, 643–650. [Google Scholar] [CrossRef]
- Cao, Y.; Su, B.; Guo, X.; Sun, W.; Deng, Y.; Bao, L.; Zhu, Q.; Zhang, X.; Zheng, Y.; Geng, C.; et al. Potent Neutralizing Antibodies against SARS-CoV-2 Identified by High-Throughput Single-Cell Sequencing of Convalescent Patients’ B Cells. Cell 2020, 182, 73–84.e16. [Google Scholar] [CrossRef]
- Chen, X.; Li, R.; Pan, Z.; Qian, C.; Yang, Y.; You, R.; Zhao, J.; Liu, P.; Gao, L.; Li, Z.; et al. Human monoclonal antibodies block the binding of SARS-CoV-2 spike protein to angiotensin converting enzyme 2 receptor. Cell Mol. Immunol. 2020, 17, 647–649. [Google Scholar] [CrossRef]
- Noy-Porat, T.; Makdasi, E.; Alcalay, R.; Mechaly, A.; Levy, Y.; Bercovich-Kinori, A.; Zauberman, A.; Tamir, H.; Yahalom-Ronen, Y.; Israeli, M.; et al. A panel of human neutralizing mAbs targeting SARS-CoV-2 spike at multiple epitopes. Nat. Commun. 2020, 11, 4303. [Google Scholar] [CrossRef]
- Shi, R.; Shan, C.; Duan, X.; Chen, Z.; Liu, P.; Song, J.; Song, T.; Bi, X.; Han, C.; Wu, L.; et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature 2020, 584, 120–124. [Google Scholar] [CrossRef]
- Chi, X.; Yan, R.; Zhang, J.; Zhang, G.; Zhang, Y.; Hao, M.; Zhang, Z.; Fan, P.; Dong, Y.; Yang, Y.; et al. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science 2020, 369, 650–655. [Google Scholar] [CrossRef]
- Liu, L.; Wang, P.; Nair, M.S.; Yu, J.; Rapp, M.; Wang, Q.; Luo, Y.; Chan, J.F.; Sahi, V.; Figueroa, A.; et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature 2020, 584, 450–456. [Google Scholar] [CrossRef]
- McCallum, M.; Marco, A.; Lempp, F.; Tortorici, M.A.; Pinto, D.; Walls, A.C.; Beltramello, M.; Chen, A.; Liu, Z.; Zatta, F.; et al. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. bioRxiv 2021. [Google Scholar] [CrossRef]
- Andreano, E.; Nicastri, E.; Paciello, I.; Pileri, P.; Manganaro, N.; Piccini, G.; Manenti, A.; Pantano, E.; Kabanova, A.; Troisi, M.; et al. Extremely potent human monoclonal antibodies from convalescent Covid-19 patients. bioRxiv 2020, 328302. [Google Scholar] [CrossRef]
- Andreano, E.; Piccini, G.; Licastro, D.; Casalino, L.; Johnson, N.V.; Paciello, I.; Monego, S.D.; Pantano, E.; Manganaro, N.; Manenti, A.; et al. SARS-CoV-2 escape in vitro from a highly neutralizing COVID-19 convalescent plasma. bioRxiv 2020, 424451. [Google Scholar] [CrossRef]
- Wang, C.; Li, W.; Drabek, D.; Okba, N.M.A.; van Haperen, R.; Osterhaus, A.; van Kuppeveld, F.J.M.; Haagmans, B.L.; Grosveld, F.; Bosch, B.J. A human monoclonal antibody blocking SARS-CoV-2 infection. Nat. Commun. 2020, 11, 2251. [Google Scholar] [CrossRef] [PubMed]
- Wrapp, D.; De Vlieger, D.; Corbett, K.S.; Torres, G.M.; Wang, N.; Van Breedam, W.; Roose, K.; van Schie, L.; Team, V.-C.C.-R.; Hoffmann, M.; et al. Structural Basis for Potent Neutralization of Betacoronaviruses by Single-Domain Camelid Antibodies. Cell 2020, 181, 1004–1015.e5. [Google Scholar] [CrossRef]
- Chan, A.C.; Carter, P.J. Therapeutic antibodies for autoimmunity and inflammation. Nat. Rev. Immunol. 2010, 10, 301–316. [Google Scholar] [CrossRef]
- Scott, A.M.; Wolchok, J.D.; Old, L.J. Antibody therapy of cancer. Nat. Rev. Cancer 2012, 12, 278–287. [Google Scholar] [CrossRef]
- Ravichandran, S.; Coyle, E.M.; Klenow, L.; Tang, J.; Grubbs, G.; Liu, S.; Wang, T.; Golding, H.; Khurana, S. Antibody signature induced by SARS-CoV-2 spike protein immunogens in rabbits. Sci. Transl. Med. 2020, 12. [Google Scholar] [CrossRef]
- Mechaly, A.; Alcalay, R.; Noy-Porat, T.; Epstein, E.; Gal, Y.; Mazor, O. Novel Phage Display-Derived Anti-Abrin Antibodies Confer Post-Exposure Protection against Abrin Intoxication. Toxins 2018, 10, 80. [Google Scholar] [CrossRef] [Green Version]
- Mechaly, A.; Elia, U.; Alcalay, R.; Cohen, H.; Epstein, E.; Cohen, O.; Mazor, O. Inhibition of Francisella tularensis phagocytosis using a novel anti-LPS scFv antibody fragment. Sci. Rep. 2019, 9, 11418. [Google Scholar] [CrossRef] [Green Version]
- Stadlbauer, D.; Amanat, F.; Chromikova, V.; Jiang, K.; Strohmeier, S.; Arunkumar, G.A.; Tan, J.; Bhavsar, D.; Capuano, C.; Kirkpatrick, E.; et al. SARS-CoV-2 Seroconversion in Humans: A Detailed Protocol for a Serological Assay, Antigen Production, and Test Setup. Curr. Protoc. Microbiol. 2020, 57, e100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020, 367, 1260–1263. [Google Scholar] [CrossRef] [Green Version]
- Noy-Porat, T.; Cohen, O.; Ehrlich, S.; Epstein, E.; Alcalay, R.; Mazor, O. Acetylcholinesterase-Fc Fusion Protein (AChE-Fc): A Novel Potential Organophosphate Bioscavenger with Extended Plasma Half-Life. Bioconjug Chem. 2015, 26, 1753–1758. [Google Scholar] [CrossRef]
- Seeber, S.; Ros, F.; Thorey, I.; Tiefenthaler, G.; Kaluza, K.; Lifke, V.; Fischer, J.A.; Klostermann, S.; Endl, J.; Kopetzki, E.; et al. A robust high throughput platform to generate functional recombinant monoclonal antibodies using rabbit B cells from peripheral blood. PLoS ONE 2014, 9, e86184. [Google Scholar] [CrossRef] [Green Version]
- Yahalom-Ronen, Y.; Tamir, H.; Melamed, S.; Politi, B.; Shifman, O.; Achdout, H.; Vitner, E.B.; Israeli, O.; Milrot, E.; Stein, D.; et al. A single dose of recombinant VSV-G-spike vaccine provides protection against SARS-CoV-2 challenge. Nat. Commun. 2020, 11, 6402. [Google Scholar] [CrossRef]
- Hansen, J.; Baum, A.; Pascal, K.E.; Russo, V.; Giordano, S.; Wloga, E.; Fulton, B.O.; Yan, Y.; Koon, K.; Patel, K.; et al. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science 2020, 369, 1010–1014. [Google Scholar] [CrossRef] [PubMed]
- Pinto, D.; Park, Y.J.; Beltramello, M.; Walls, A.C.; Tortorici, M.A.; Bianchi, S.; Jaconi, S.; Culap, K.; Zatta, F.; De Marco, A.; et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature 2020, 583, 290–295. [Google Scholar] [CrossRef]
- Renn, A.; Fu, Y.; Hu, X.; Hall, M.D.; Simeonov, A. Fruitful Neutralizing Antibody Pipeline Brings Hope To Defeat SARS-Cov-2. Trends Pharmacol. Sci. 2020, 41, 815–829. [Google Scholar] [CrossRef]
- He, Y.; Zhou, Y.; Liu, S.; Kou, Z.; Li, W.; Farzan, M.; Jiang, S. Receptor-binding domain of SARS-CoV spike protein induces highly potent neutralizing antibodies: Implication for developing subunit vaccine. Biochem. Biophys Res. Commun. 2004, 324, 773–781. [Google Scholar] [CrossRef] [PubMed]
- Ren, W.; Sun, H.; Gao, G.F.; Chen, J.; Sun, S.; Zhao, R.; Gao, G.; Hu, Y.; Zhao, G.; Chen, Y.; et al. Recombinant SARS-CoV-2 spike S1-Fc fusion protein induced high levels of neutralizing responses in nonhuman primates. Vaccine 2020, 38, 5653–5658. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, L.; Cao, H.; Liu, C. SARS-CoV-2 S1 is superior to the RBD as a COVID-19 subunit vaccine antigen. J. Med. Virol. 2021, 93, 892–898. [Google Scholar] [CrossRef] [PubMed]
- Wan, J.; Xing, S.; Ding, L.; Wang, Y.; Gu, C.; Wu, Y.; Rong, B.; Li, C.; Wang, S.; Chen, K.; et al. Human-IgG-Neutralizing Monoclonal Antibodies Block the SARS-CoV-2 Infection. Cell Rep. 2020, 32, 107918. [Google Scholar] [CrossRef]
- Lv, Z.; Deng, Y.Q.; Ye, Q.; Cao, L.; Sun, C.Y.; Fan, C.; Huang, W.; Sun, S.; Sun, Y.; Zhu, L.; et al. Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody. Science 2020, 369, 1505–1509. [Google Scholar] [CrossRef] [PubMed]
- Baisa, G.; Rancour, D.; Mansfield, K.; Burns, M.; Martin, L.; Cunha, D.; Fischer, J.; Muecksch, F.; Hatziioannou, T.; Bieniasz, P.; et al. A recombinant protein SARS-CoV-2 candidate vaccine elicits high-titer neutralizing antibodies in macaques. bioRxiv 2020, 422693. [Google Scholar] [CrossRef]
- Diamond, M.; Chen, R.; Xie, X.; Case, J.; Zhang, X.; VanBlargan, L.; Liu, Y.; Liu, J.; Errico, J.; Winkler, E.; et al. SARS-CoV-2 variants show resistance to neutralization by many monoclonal and serum-derived polyclonal antibodies. Res. Sq 2021. [Google Scholar] [CrossRef]
- Galloway, S.E.; Paul, P.; MacCannell, D.R.; Johansson, M.A.; Brooks, J.T.; MacNeil, A.; Slayton, R.B.; Tong, S.; Silk, B.J.; Armstrong, G.L.; et al. Emergence of SARS-CoV-2 B.1.1.7 Lineage-United States, December 29, 2020-January 12, 2021. MMWR Morb Mortal. Wkly. Rep. 2021, 70, 95–99. [Google Scholar] [CrossRef]
- Greaney, A.J.; Starr, T.N.; Gilchuk, P.; Zost, S.J.; Binshtein, E.; Loes, A.N.; Hilton, S.K.; Huddleston, J.; Eguia, R.; Crawford, K.H.D.; et al. Complete Mapping of Mutations to the SARS-CoV-2 Spike Receptor-Binding Domain that Escape Antibody Recognition. Cell Host Microbe 2021, 29, 44–57.e49. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; VanBlargan, L.A.; Bloyet, L.M.; Rothlauf, P.W.; Chen, R.E.; Stumpf, S.; Zhao, H.; Errico, J.M.; Theel, E.S.; Liebeskind, M.J.; et al. Identification of SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. Cell Host Microbe 2021. [Google Scholar] [CrossRef]
- Tegally, H.; Wilkinson, E.; Lessells, R.J.; Giandhari, J.; Pillay, S.; Msomi, N.; Mlisana, K.; Bhiman, J.N.; von Gottberg, A.; Walaza, S.; et al. Sixteen novel lineages of SARS-CoV-2 in South Africa. Nat. Med. 2021. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Makdasi, E.; Levy, Y.; Alcalay, R.; Noy-Porat, T.; Zahavy, E.; Mechaly, A.; Epstein, E.; Peretz, E.; Cohen, H.; Bar-On, L.; et al. Neutralizing Monoclonal Anti-SARS-CoV-2 Antibodies Isolated from Immunized Rabbits Define Novel Vulnerable Spike-Protein Epitope. Viruses 2021, 13, 566. https://doi.org/10.3390/v13040566
Makdasi E, Levy Y, Alcalay R, Noy-Porat T, Zahavy E, Mechaly A, Epstein E, Peretz E, Cohen H, Bar-On L, et al. Neutralizing Monoclonal Anti-SARS-CoV-2 Antibodies Isolated from Immunized Rabbits Define Novel Vulnerable Spike-Protein Epitope. Viruses. 2021; 13(4):566. https://doi.org/10.3390/v13040566
Chicago/Turabian StyleMakdasi, Efi, Yinon Levy, Ron Alcalay, Tal Noy-Porat, Eran Zahavy, Adva Mechaly, Eyal Epstein, Eldar Peretz, Hila Cohen, Liat Bar-On, and et al. 2021. "Neutralizing Monoclonal Anti-SARS-CoV-2 Antibodies Isolated from Immunized Rabbits Define Novel Vulnerable Spike-Protein Epitope" Viruses 13, no. 4: 566. https://doi.org/10.3390/v13040566





