Host Cell Entry and Neutralization Sensitivity of SARS-CoV-2 Lineages B.1.620 and R.1
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Plasmids
2.3. Production of Pseudotype Particles
2.4. Analysis of Spike Protein-Mediated Cell Entry
2.5. Production of Soluble ACE2
2.6. Analysis of ACE2 Binding by Flow Cytometry
2.7. Collection of Serum and Plasma Samples
2.8. Neutralization Assay
2.9. Data Analysis
3. Results
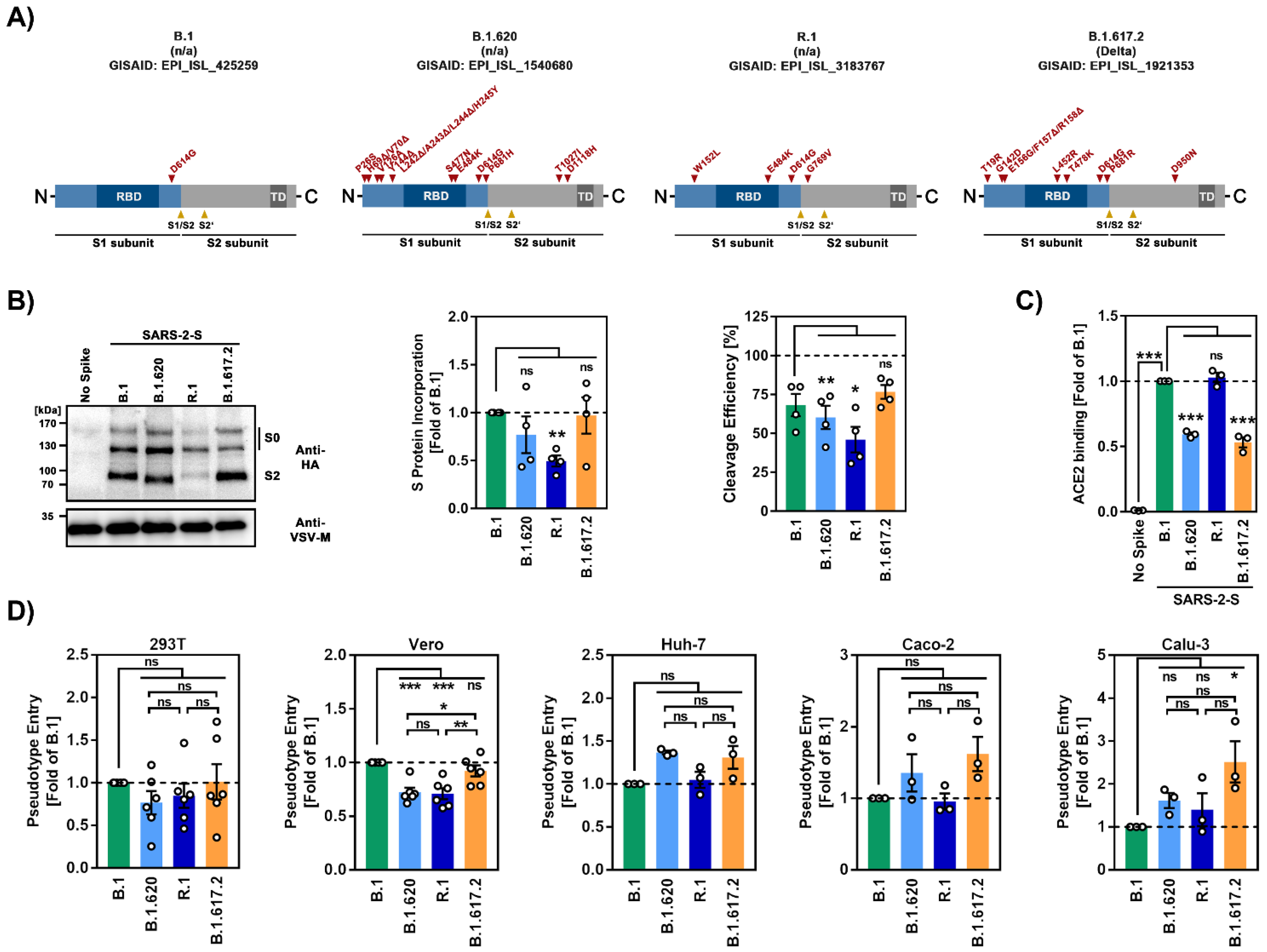
3.1. The Spike Proteins of SARS-CoV-2 Lineages B.1.620 and R.1 Differ Greatly with Respect to the Number of Mutations
3.2. The Spike Proteins of SARS-CoV-2 Lineages B.1.620 and R.1 Differ Regarding Cleavability, Particle Incorporation and ACE2 Binding Compared to the Spike Protein of the SARS-CoV-2 B.1 Lineage
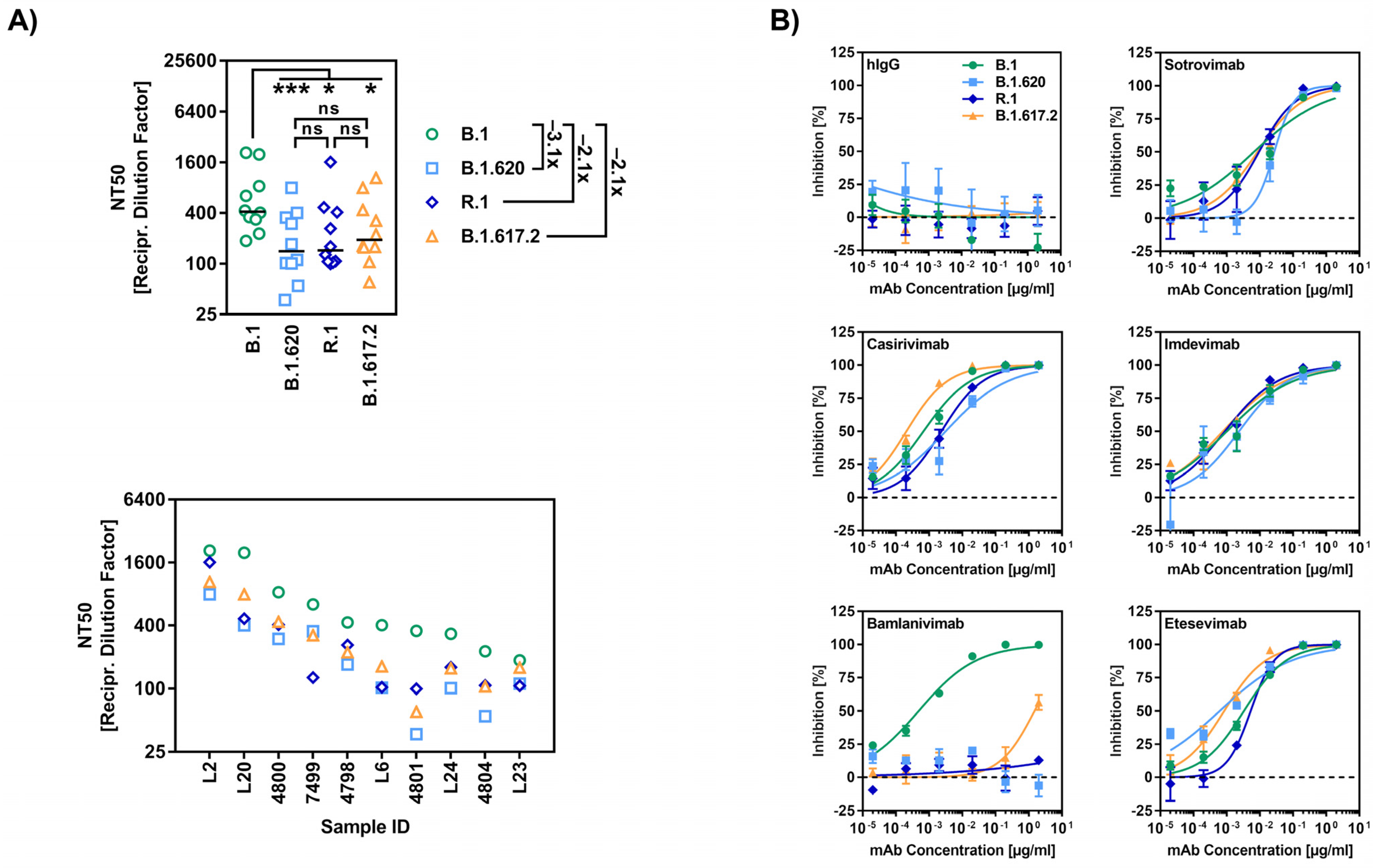
3.3. The Spike Proteins of SARS-CoV-2 Lineages B.1.620 and R.1 Display Reduced Sensitivity to Neutralization by Antibodies Induced upon Vaccination and Clinically-Used Monoclonal Antibodies
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Favresse, J.; Gillot, C.; Di Chiaro, L.; Eucher, C.; Elsen, M.; Van Eeckhoudt, S.; David, C.; Morimont, L.; Dogné, J.-M.; Douxfils, J. Neutralizing Antibodies in COVID-19 Patients and Vaccine Recipients after Two Doses of BNT162b2. Viruses 2021, 13, 1364. [Google Scholar] [CrossRef] [PubMed]
- Baden, L.R.; El Sahly, H.M.; Essink, B.; Kotloff, K.; Frey, S.; Novak, R.; Diemert, D.; Spector, S.A.; Rouphael, N.; Creech, C.B.; et al. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med. 2020, 384, 403–416. [Google Scholar] [CrossRef] [PubMed]
- Polack, F.P.; Thomas, S.J.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Pérez Marc, G.; Moreira, E.D.; Zerbini, C.; et al. Safety and Efficacy of the BNT162b2 mRNA COVID-19 Vaccine. N. Engl. J. Med. 2020, 383, 2603–2615. [Google Scholar] [CrossRef] [PubMed]
- Baum, A.; Ajithdoss, D.; Copin, R.; Zhou, A.; Lanza, K.; Negron, N.; Ni, M.; Wei, Y.; Mohammadi, K.; Musser, B.; et al. REGN-COV2 antibodies prevent and treat SARS-CoV-2 infection in rhesus macaques and hamsters. Science 2020, 370, 1110–1115. [Google Scholar] [CrossRef]
- Chen, P.; Nirula, A.; Heller, B.; Gottlieb, R.L.; Boscia, J.; Morris, J.; Huhn, G.; Cardona, J.; Mocherla, B.; Stosor, V.; et al. SARS-CoV-2 Neutralizing Antibody LY-CoV555 in Outpatients with COVID-19. N. Engl. J. Med. 2020, 384, 229–237. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Kruger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e278. [Google Scholar] [CrossRef]
- Zhou, T.; Tsybovsky, Y.; Gorman, J.; Rapp, M.; Cerutti, G.; Chuang, G.-Y.; Katsamba, P.S.; Sampson, J.M.; Schön, A.; Bimela, J.; et al. Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains. Cell Host Microbe 2020, 28, 867–879.e865. [Google Scholar] [CrossRef]
- Yang, Y.; Du, L. SARS-CoV-2 spike protein: A key target for eliciting persistent neutralizing antibodies. Signal Transduct. Target. Ther. 2021, 6, 95. [Google Scholar] [CrossRef]
- Brinkmann, C.; Hoffmann, M.; Lubke, A.; Nehlmeier, I.; Kramer-Kuhl, A.; Winkler, M.; Pohlmann, S. The glycoprotein of vesicular stomatitis virus promotes release of virus-like particles from tetherin-positive cells. PLoS ONE 2017, 12, e0189073. [Google Scholar] [CrossRef] [Green Version]
- Hoffmann, M.; Arora, P.; Gross, R.; Seidel, A.; Hornich, B.F.; Hahn, A.S.; Kruger, N.; Graichen, L.; Hofmann-Winkler, H.; Kempf, A.; et al. SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell 2021, 184, 2384–2393.e2312. [Google Scholar] [CrossRef]
- Arora, P.; Sidarovich, A.; Kruger, N.; Kempf, A.; Nehlmeier, I.; Graichen, L.; Moldenhauer, A.S.; Winkler, M.S.; Schulz, S.; Jack, H.M.; et al. B.1.617.2 enters and fuses lung cells with increased efficiency and evades antibodies induced by infection and vaccination. Cell Rep. 2021, 37, 109825. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Kruger, N.; Schulz, S.; Cossmann, A.; Rocha, C.; Kempf, A.; Nehlmeier, I.; Graichen, L.; Moldenhauer, A.S.; Winkler, M.S.; et al. The Omicron variant is highly resistant against antibody-mediated neutralization: Implications for control of the COVID-19 pandemic. Cell 2022, 185, 447–456.e411. [Google Scholar] [CrossRef] [PubMed]
- Kleine-Weber, H.; Elzayat, M.T.; Wang, L.; Graham, B.S.; Muller, M.A.; Drosten, C.; Pohlmann, S.; Hoffmann, M. Mutations in the Spike Protein of Middle East Respiratory Syndrome Coronavirus Transmitted in Korea Increase Resistance to Antibody-Mediated Neutralization. J. Virol. 2019, 93, e01381-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berger Rentsch, M.; Zimmer, G. A vesicular stomatitis virus replicon-based bioassay for the rapid and sensitive determination of multi-species type I interferon. PLoS ONE 2011, 6, e25858. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoffmann, M.; Zhang, L.; Kruger, N.; Graichen, L.; Kleine-Weber, H.; Hofmann-Winkler, H.; Kempf, A.; Nessler, S.; Riggert, J.; Winkler, M.S.; et al. SARS-CoV-2 mutations acquired in mink reduce antibody-mediated neutralization. Cell Rep. 2021, 35, 109017. [Google Scholar] [CrossRef] [PubMed]
- Dudas, G.; Hong, S.L.; Potter, B.I.; Calvignac-Spencer, S.; Niatou-Singa, F.S.; Tombolomako, T.B.; Fuh-Neba, T.; Vickos, U.; Ulrich, M.; Leendertz, F.H.; et al. Emergence and spread of SARS-CoV-2 lineage B.1.620 with variant of concern-like mutations and deletions. Nat. Commun. 2021, 12, 5769. [Google Scholar] [CrossRef]
- Chi, X.; Yan, R.; Zhang, J.; Zhang, G.; Zhang, Y.; Hao, M.; Zhang, Z.; Fan, P.; Dong, Y.; Yang, Y.; et al. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science 2020, 369, 650–655. [Google Scholar] [CrossRef]
- Liu, L.; Wang, P.; Nair, M.S.; Yu, J.; Rapp, M.; Wang, Q.; Luo, Y.; Chan, J.F.W.; Sahi, V.; Figueroa, A.; et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature 2020, 584, 450–456. [Google Scholar] [CrossRef]
- McCallum, M.; De Marco, A.; Lempp, F.A.; Tortorici, M.A.; Pinto, D.; Walls, A.C.; Beltramello, M.; Chen, A.; Liu, Z.; Zatta, F.; et al. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. Cell 2021, 184, 2332–2347.e2316. [Google Scholar] [CrossRef]
- Suryadevara, N.; Shrihari, S.; Gilchuk, P.; VanBlargan, L.A.; Binshtein, E.; Zost, S.J.; Nargi, R.S.; Sutton, R.E.; Winkler, E.S.; Chen, E.C.; et al. Neutralizing and protective human monoclonal antibodies recognizing the N-terminal domain of the SARS-CoV-2 spike protein. Cell 2021, 184, 2316–2331.e2315. [Google Scholar] [CrossRef]
- Singh, A.; Steinkellner, G.; Köchl, K.; Gruber, K.; Gruber, C.C. Serine 477 plays a crucial role in the interaction of the SARS-CoV-2 spike protein with the human receptor ACE2. Sci. Rep. 2021, 11, 4320. [Google Scholar] [CrossRef] [PubMed]
- Mejdani, M.; Haddadi, K.; Pham, C.; Mahadevan, R. SARS-CoV-2 receptor-binding mutations and antibody contact sites. Antib. Ther. 2021, 4, 149–158. [Google Scholar] [CrossRef] [PubMed]
- Jangra, S.; Ye, C.; Rathnasinghe, R.; Stadlbauer, D.; Alshammary, H.; Amoako, A.A.; Awawda, M.H.; Beach, K.F.; Bermúdez-González, M.C.; Chernet, R.L.; et al. SARS-CoV-2 spike E484K mutation reduces antibody neutralisation. Lancet Microbe 2021, 2, e283–e284. [Google Scholar] [CrossRef]
- Jangra, S.; Ye, C.; Rathnasinghe, R.; Stadlbauer, D.; group, P.V.I.s.; Krammer, F.; Simon, V.; Martinez-Sobrido, L.; García-Sastre, A.; Schotsaert, M. The E484K mutation in the SARS-CoV-2 spike protein reduces but does not abolish neutralizing activity of human convalescent and post-vaccination sera. medRxiv 2021, arXiv:2021.2001.2026.21250543. [Google Scholar] [CrossRef]
- Lassaunière, R.; Polacek, C.; Fonager, J.; Bennedbæk, M.; Boding, L.; Rasmussen, M.; Fomsgaard, A. Neutralisation of the SARS-CoV-2 Delta sub-lineage AY.4.2 and B.1.617.2 + E484K by BNT162b2 mRNA vaccine-elicited sera. medRxiv 2021, arXiv:2021.2011.2008.21266075. [Google Scholar] [CrossRef]
- Chen, R.E.; Zhang, X.; Case, J.B.; Winkler, E.S.; Liu, Y.; VanBlargan, L.A.; Liu, J.; Errico, J.M.; Xie, X.; Suryadevara, N.; et al. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat. Med. 2021, 27, 717–726. [Google Scholar] [CrossRef]
- Plante, J.A.; Liu, Y.; Liu, J.; Xia, H.; Johnson, B.A.; Lokugamage, K.G.; Zhang, X.; Muruato, A.E.; Zou, J.; Fontes-Garfias, C.R.; et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature 2021, 592, 116–121. [Google Scholar] [CrossRef]
- Zhou, B.; Thao, T.T.N.; Hoffmann, D.; Taddeo, A.; Ebert, N.; Labroussaa, F.; Pohlmann, A.; King, J.; Steiner, S.; Kelly, J.N.; et al. SARS-CoV-2 spike D614G change enhances replication and transmission. Nature 2021, 592, 122–127. [Google Scholar] [CrossRef]
- Lubinski, B.; Fernandes, M.H.V.; Frazier, L.; Tang, T.; Daniel, S.; Diel, D.G.; Jaimes, J.A.; Whittaker, G.R. Functional evaluation of the P681H mutation on the proteolytic activation of the SARS-CoV-2 variant B.1.1.7 (Alpha) spike. iScience 2022, 25, 103589. [Google Scholar] [CrossRef]
- Arora, P.; Sidarovich, A.; Graichen, L.; Hornich, B.; Hahn, A.; Hoffmann, M.; Pohlmann, S. Functional analysis of polymorphisms at the S1/S2 site of SARS-CoV-2 spike protein. PLoS ONE 2022, 17, e0265453. [Google Scholar] [CrossRef]
- Hirotsu, Y.; Omata, M. Detection of R.1 lineage severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) with spike protein W152L/E484K/G769V mutations in Japan. PLoS Pathog. 2021, 17, e1009619. [Google Scholar] [CrossRef] [PubMed]
- Hirotsu, Y.; Omata, M. SARS-CoV-2 B.1.1.7 lineage rapidly spreads and replaces R.1 lineage in Japan: Serial and stationary observation in a community. Infect. Genet. Evol. 2021, 95, 105088. [Google Scholar] [CrossRef] [PubMed]
- Sekizuka, T.; Itokawa, K.; Hashino, M.; Okubo, K.; Ohnishi, A.; Goto, K.; Tsukagoshi, H.; Ehara, H.; Nomoto, R.; Ohnishi, M.; et al. A discernable increase in the severe acute respiratory syndrome coronavirus 2 R.1 lineage carrying an E484K spike protein mutation in Japan. Infect. Genet. Evol. 2021, 94, 105013. [Google Scholar] [CrossRef] [PubMed]
- Nagano, K.; Tani-Sassa, C.; Iwasaki, Y.; Takatsuki, Y.; Yuasa, S.; Takahashi, Y.; Nakajima, J.; Sonobe, K.; Ichimura, N.; Nukui, Y.; et al. SARS-CoV-2 R.1 lineage variants that prevailed in Tokyo in March 2021. J. Med. Virol. 2021, 93, 6833–6836. [Google Scholar] [CrossRef]
- Cavanaugh, A.M.; Fortier, S.; Lewis, P.; Arora, V.; Johnson, M.; George, K.; Tobias, J.; Lunn, S.; Miller, T.; Thoroughman, D.; et al. COVID-19 Outbreak Associated with a SARS-CoV-2 R.1 Lineage Variant in a Skilled Nursing Facility After Vaccination Program—Kentucky, March 2021. MMWR Morb. Mortal Wkly. Rep. 2021, 70, 639–643. [Google Scholar] [CrossRef]
- Ip, J.D.; Kok, K.H.; Chan, W.M.; Chu, A.W.; Wu, W.L.; Yip, C.C.; To, W.K.; Tsang, O.T.; Leung, W.S.; Chik, T.S.; et al. Intra-host non-synonymous diversity at a neutralizing antibody epitope of SARS-CoV-2 spike protein N-terminal domain. Clin. Microbiol. Infect. 2021, 27, 1350.e1351–1350.e1355. [Google Scholar] [CrossRef]
- Schmidt, F.; Weisblum, Y.; Muecksch, F.; Hoffmann, H.H.; Michailidis, E.; Lorenzi, J.C.C.; Mendoza, P.; Rutkowska, M.; Bednarski, E.; Gaebler, C.; et al. Measuring SARS-CoV-2 neutralizing antibody activity using pseudotyped and chimeric viruses. J. Exp. Med. 2020, 217. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, J.; Johnson, B.A.; Xia, H.; Ku, Z.; Schindewolf, C.; Widen, S.G.; An, Z.; Weaver, S.C.; Menachery, V.D.; et al. Delta spike P681R mutation enhances SARS-CoV-2 fitness over Alpha variant. Cell Rep. 2022, 39, 110829. [Google Scholar] [CrossRef]
- Saito, A.; Irie, T.; Suzuki, R.; Maemura, T.; Nasser, H.; Uriu, K.; Kosugi, Y.; Shirakawa, K.; Sadamasu, K.; Kimura, I.; et al. Enhanced fusogenicity and pathogenicity of SARS-CoV-2 Delta P681R mutation. Nature 2022, 602, 300–306. [Google Scholar] [CrossRef]
- Arora, P.; Kempf, A.; Nehlmeier, I.; Graichen, L.; Sidarovich, A.; Winkler, M.S.; Schulz, S.; Jack, H.M.; Stankov, M.V.; Behrens, G.M.N.; et al. Delta variant (B.1.617.2) sublineages do not show increased neutralization resistance. Cell. Mol. Immunol. 2021, 18, 2557–2559. [Google Scholar] [CrossRef]
- Mlcochova, P.; Kemp, S.A.; Dhar, M.S.; Papa, G.; Meng, B.; Ferreira, I.; Datir, R.; Collier, D.A.; Albecka, A.; Singh, S.; et al. SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion. Nature 2021, 599, 114–119. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Sidarovich, A.; Arora, P.; Kruger, N.; Nehlmeier, I.; Kempf, A.; Graichen, L.; Winkler, M.S.; Niemeyer, D.; Goffinet, C.; et al. Evidence for an ACE2-Independent Entry Pathway That Can Protect from Neutralization by an Antibody Used for COVID-19 Therapy. mBio 2022, 13, e0036422. [Google Scholar] [CrossRef] [PubMed]
- Yuan, M.; Huang, D.; Lee, C.D.; Wu, N.C.; Jackson, A.M.; Zhu, X.; Liu, H.; Peng, L.; van Gils, M.J.; Sanders, R.W.; et al. Structural and functional ramifications of antigenic drift in recent SARS-CoV-2 variants. Science 2021, 373, 818–823. [Google Scholar] [CrossRef] [PubMed]
- Mannar, D.; Saville, J.W.; Zhu, X.; Srivastava, S.S.; Berezuk, A.M.; Tuttle, K.S.; Marquez, A.C.; Sekirov, I.; Subramaniam, S. SARS-CoV-2 Omicron variant: Antibody evasion and cryo-EM structure of spike protein-ACE2 complex. Science 2022, 375, 760–764. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Jackson, C.B.; Mou, H.; Ojha, A.; Peng, H.; Quinlan, B.D.; Rangarajan, E.S.; Pan, A.; Vanderheiden, A.; Suthar, M.S.; et al. SARS-CoV-2 spike-protein D614G mutation increases virion spike density and infectivity. Nat. Commun. 2020, 11, 6013. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Pohlmann, S. A Multibasic Cleavage Site in the Spike Protein of SARS-CoV-2 Is Essential for Infection of Human Lung Cells. Mol. Cell 2020, 78, 779–784.e775. [Google Scholar] [CrossRef]
- Starr, T.N.; Greaney, A.J.; Dingens, A.S.; Bloom, J.D. Complete map of SARS-CoV-2 RBD mutations that escape the monoclonal antibody LY-CoV555 and its cocktail with LY-CoV016. Cell Rep. Med. 2021, 2, 100255. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sidarovich, A.; Krüger, N.; Rocha, C.; Graichen, L.; Kempf, A.; Nehlmeier, I.; Lier, M.; Cossmann, A.; Stankov, M.V.; Schulz, S.R.; et al. Host Cell Entry and Neutralization Sensitivity of SARS-CoV-2 Lineages B.1.620 and R.1. Viruses 2022, 14, 2475. https://doi.org/10.3390/v14112475
Sidarovich A, Krüger N, Rocha C, Graichen L, Kempf A, Nehlmeier I, Lier M, Cossmann A, Stankov MV, Schulz SR, et al. Host Cell Entry and Neutralization Sensitivity of SARS-CoV-2 Lineages B.1.620 and R.1. Viruses. 2022; 14(11):2475. https://doi.org/10.3390/v14112475
Chicago/Turabian StyleSidarovich, Anzhalika, Nadine Krüger, Cheila Rocha, Luise Graichen, Amy Kempf, Inga Nehlmeier, Martin Lier, Anne Cossmann, Metodi V. Stankov, Sebastian R. Schulz, and et al. 2022. "Host Cell Entry and Neutralization Sensitivity of SARS-CoV-2 Lineages B.1.620 and R.1" Viruses 14, no. 11: 2475. https://doi.org/10.3390/v14112475




