Rabies Virus Populations in Humans and Mice Show Minor Inter-Host Variability within Various Central Nervous System Regions and Peripheral Tissues
Abstract
:1. Introduction
2. Materials and Methods
2.1. Human Materials and Cell Culture
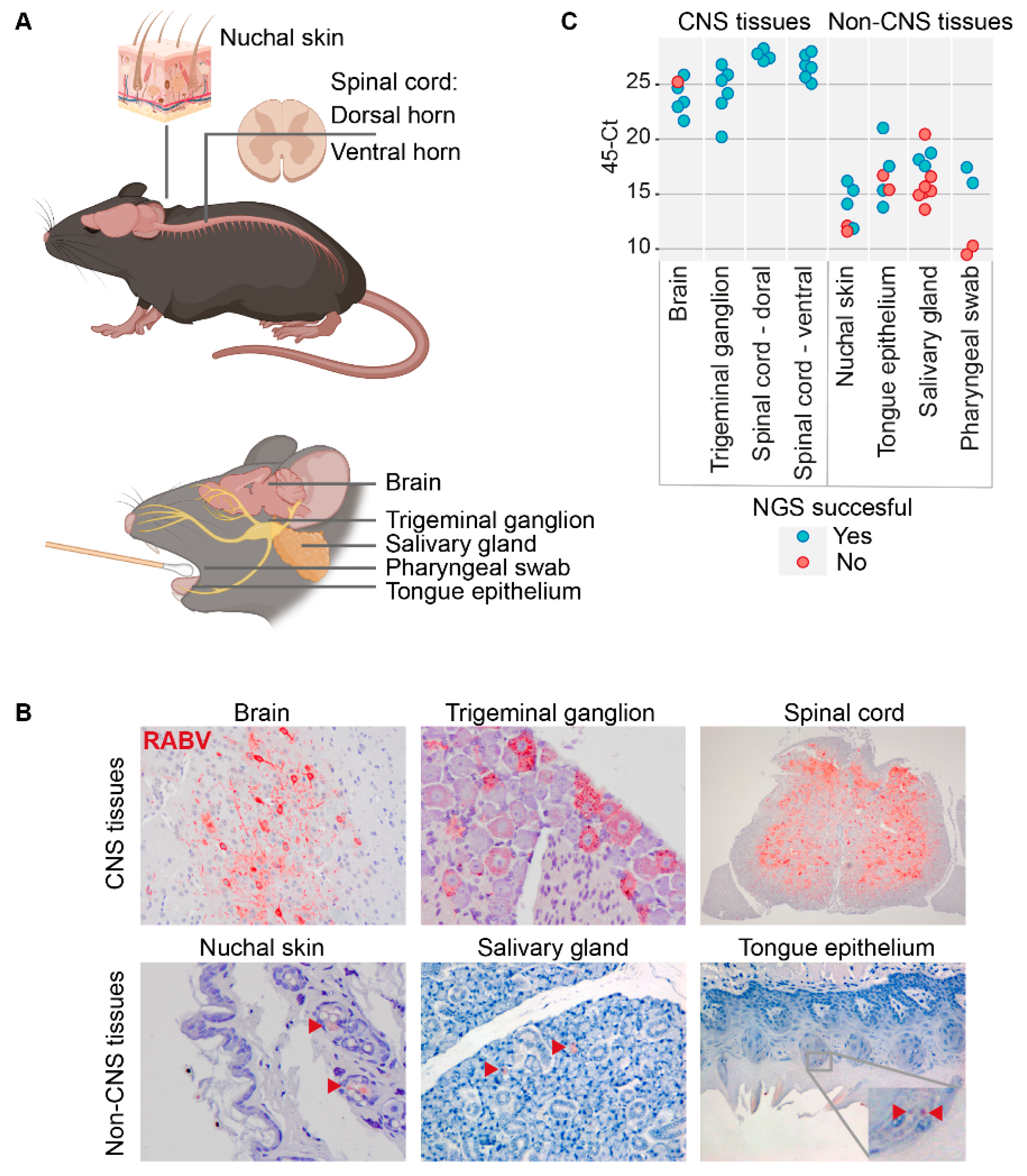
2.2. Animal Materials
2.3. Sample Preparation and RNA Isolation
2.4. cDNA Library Preparation and Sequencing
2.5. Minor Variant Analysis
2.6. Immunohistochemistry and Immunofluorescence
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO. Expert Consultation on Rabies-Third Report; WHO: Geneva, Switzerland, 2018. [Google Scholar]
- Hampson, K.; Coudeville, L.; Lembo, T.; Sambo, M.; Kieffer, A.; Attlan, M.; Barrat, J.; Blanton, J.D.; Briggs, D.J.; Cleaveland, S.; et al. Estimating the Global Burden of Endemic Canine Rabies. PLoS Negl. Trop. Dis. 2015, 9, e0003709. [Google Scholar] [CrossRef] [Green Version]
- Taylor, L.H.; Hampson, K.; Fahrion, A.; Abela-Ridder, B.; Nel, L.H. Difficulties in Estimating the Human Burden of Canine Rabies. Acta Trop. 2017, 165, 133–140. [Google Scholar] [CrossRef] [PubMed]
- Benavides, J.A.; Rojas Paniagua, E.; Hampson, K.; Valderrama, W.; Streicker, D.G. Quantifying the Burden of Vampire Bat Rabies in Peruvian Livestock. PLoS Negl. Trop. Dis. 2017, 11, e0006105. [Google Scholar] [CrossRef] [Green Version]
- Charlton, K.M.; Casey, G.A. Experimental Rabies in Skunks: Immunofluorescence Light and Electron Microscopic Studies. Lab. Investig. 1979, 41, 36–44. [Google Scholar] [PubMed]
- Allendorf, S.D.; Cortez, A.; Heinemann, M.B.; Harary, C.M.A.; Antunes, J.M.A.; Peres, M.G.; Vicente, A.F.; Sodré, M.M.; da Rosa, A.R.; Megid, J. Rabies Virus Distribution in Tissues and Molecular Characterization of Strains from Naturally Infected Non-Hematophagous Bats. Virus Res. 2012, 165, 119–125. [Google Scholar] [CrossRef] [Green Version]
- Delmas, O.; Holmes, E.C.; Talbi, C.; Larrous, F.; Dacheux, L.; Bouchier, C.; Bourhy, H. Genomic Diversity and Evolution of the Lyssaviruses. PLoS ONE 2008, 3, e2057. [Google Scholar] [CrossRef] [PubMed]
- Troupin, C.; Dacheux, L.; Tanguy, M.; Sabeta, C.; Blanc, H.; Bouchier, C.; Vignuzzi, M.; Duchene, S.; Holmes, E.C.; Bourhy, H. Large-Scale Phylogenomic Analysis Reveals the Complex Evolutionary History of Rabies Virus in Multiple Carnivore Hosts. PLoS Pathog. 2016, 12, e1006041. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sooksawasdi Na Ayudhya, S.; Meijer, A.; Bauer, L.; Oude Munnink, B.; Embregts, C.; Leijten, L.; Siegers, J.Y.; Laksono, B.M.; van Kuppeveld, F.; Kuiken, T.; et al. Enhanced Enterovirus D68 Replication in Neuroblastoma Cells Is Associated with a Cell Culture-Adaptive Amino Acid Substitution in VP1. Msphere 2020, 5, e00941-20. [Google Scholar] [CrossRef] [PubMed]
- Nadin-Davis, S.A.; Fehlner-Gardiner, C. Chapter 5 Lyssaviruses-Current Trends. Adv. Virus Res. 2008, 71, 207–250. [Google Scholar] [CrossRef]
- Marston, D.A.; Banyard, A.C.; McElhinney, L.M.; Freuling, C.M.; Finke, S.; de Lamballerie, X.; Müller, T.; Fooks, A.R. The Lyssavirus Host-Specificity Conundrum—Rabies Virus—the Exception Not the Rule. Curr. Opin. Virol. 2018, 28, 68–73. [Google Scholar] [CrossRef]
- Munnink, B.B.O.; Abd Farag, E.A.B.; GeurtsvanKessel, C.; Schapendonk, C.; van der Linden, A.; Kohl, R.; Arron, G.; Ziglam, H.; Goravey, W.G.M.; Coyle, P.V.; et al. First Molecular Analysis of Rabies Virus in Qatar and Clinical Cases Imported into Qatar, a Case Report. Int. J. Infect. Dis. 2020, 96, 323–326. [Google Scholar] [CrossRef] [PubMed]
- Scott, T.P.; Fischer, M.; Khaiseb, S.; Freuling, C.; Höper, D.; Hoffmann, B.; Markotter, W.; Müller, T.; Nel, L.H. Complete Genome and Molecular Epidemiological Data Infer the Maintenance of Rabies among Kudu (Tragelaphus strepsiceros) in Namibia. PLoS ONE 2013, 8, e58739. [Google Scholar] [CrossRef] [PubMed]
- Bonnaud, E.M.; Troupin, C.; Dacheux, L.; Holmes, E.C.; Monchatre-Leroy, E.; Tanguy, M.; Bouchier, C.; Cliquet, F.; Barrat, J.; Bourhy, H. Comparison of Intra- and Inter-Host Genetic Diversity in Rabies Virus during Experimental Cross-Species Transmission. PLOS Pathog. 2019, 15, e1007799. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kissi, B.; Badrane, H.; Audry, L.; Lavenu, A.; Tordo, N.; Brahimi, M.; Bourhy, H. Dynamics of Rabies Virus Quasispecies during Serial Passages in Heterologous Hosts. J. Gen. Virol. 1999, 80, 2041–2050. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holmes, E.C.; Woelk, C.H.; Kassis, R.; Bourhy, H. Genetic Constraints and the Adaptive Evolution of Rabies Virus in Nature. Virology 2002, 292, 247–257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koraka, P.; Martina, B.E.; Smreczak, M.; Orlowska, A.; Marzec, A.; Trebas, P.; Roose, J.M.; Begeman, L.; Gerhauser, I.; Wohlsein, P.; et al. Inhibition of Caspase-1 Prolongs Survival of Mice Infected with Rabies Virus. Vaccine 2019, 37, 4681–4685. [Google Scholar] [CrossRef]
- Marosi, A.; Dufkova, L.; Forró, B.; Felde, O.; Erdélyi, K.; Širmarová, J.; Palus, M.; Hönig, V.; Salát, J.; Tikos, R.; et al. Combination Therapy of Rabies-Infected Mice with Inhibitors of pro-Inflammatory Host Response, Antiviral Compounds and Human Rabies Immunoglobulin. Vaccine 2019, 37, 4724–4735. [Google Scholar] [CrossRef]
- Marston, D.A.; Horton, D.L.; Nunez, J.; Ellis, R.J.; Orton, R.J.; Johnson, N.; Banyard, A.C.; McElhinney, L.M.; Freuling, C.M.; Fırat, M.; et al. Genetic Analysis of a Rabies Virus Host Shift Event Reveals Within-Host Viral Dynamics in a New Host. Virus Evol. 2017, 3, vex038. [Google Scholar] [CrossRef] [Green Version]
- Wakeley, P.R.; Johnson, N.; McElhinney, L.M.; Marston, D.; Sawyer, J.; Fooks, A.R. Development of a Real-Time, Differential RT-PCR TaqMan® Assay for Lyssavirus Genotypes 1, 5 and 6. Dev. Biol. 2006, 126, 227–236. [Google Scholar] [CrossRef]
- Wadhwa, A.; Wilkins, K.; Gao, J.; Condori Condori, R.E.; Gigante, C.M.; Zhao, H.; Ma, X.; Ellison, J.A.; Greenberg, L.; Velasco-Villa, A.; et al. A Pan-Lyssavirus Taqman Real-Time RT-PCR Assay for the Detection of Highly Variable Rabies Virus and Other Lyssaviruses. PLoS Negl. Trop. Dis. 2017, 11, e0005258. [Google Scholar] [CrossRef]
- Briese, T.; Kapoor, A.; Mishra, N.; Jain, K.; Kumar, A.; Jabado, O.J.; Ian Lipkina, W. Virome Capture Sequencing Enables Sensitive Viral Diagnosis and Comprehensive Virome Analysis. MBio 2015, 6, e01491-15. [Google Scholar] [CrossRef] [Green Version]
- Morimoto, K.; Hooper, D.C.; Carbaugh, H.; Fu, Z.F.; Koprowski, H.; Dietzschold, B. Rabies Virus Quasispecies: Implications for Pathogenesis. Proc. Natl. Acad. Sci. USA 1998, 95, 3152–3156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watermann, C.; Valerius, K.P.; Wagner, S.; Wittekindt, C.; Klussmann, J.P.; Baumgart-Vogt, E.; Karnati, S. Step-by-Step Protocol to Perfuse and Dissect the Mouse Parotid Gland and Isolation of High-Quality RNA from Murine and Human Parotid Tissue. Biotechniques 2016, 60, 200–203. [Google Scholar] [CrossRef] [Green Version]
- Jansson, L.; Akel, Y.; Eriksson, R.; Lavander, M.; Hedman, J. Impact of Swab Material on Microbial Surface Sampling. J. Microbiol. Methods 2020, 176, 106006. [Google Scholar] [CrossRef] [PubMed]
- Roque, M.; Proudfoot, K.; Mathys, V.; Yu, S.; Krieger, N.; Gernon, T.; Gokli, K.; Hamilton, S.; Cook, C.; Fong, Y. A Review of Nasopharyngeal Swab and Saliva Tests for SARS-CoV-2 Infection: Disease Timelines, Relative Sensitivities, and Test Optimization. J. Surg. Oncol. 2021, 124, 465–475. [Google Scholar] [CrossRef]
- Segal, K.; Lisnyansky, I.; Nageris, B.; Feinmesser, R. Parasympathetic Innervation of the Salivary Glands. Oper. Tech. Otolaryngol.-Head Neck Surg. 1996, 7, 333–338. [Google Scholar] [CrossRef]
- Borucki, M.K.; Chen-Harris, H.; Lao, V.; Vanier, G.; Wadford, D.A.; Messenger, S.; Allen, J.E. Ultra-Deep Sequencing of Intra-Host Rabies Virus Populations during Cross-Species Transmission. PLoS Negl. Trop. Dis. 2013, 7, e2555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Novaes Oliveira, R.; de Souza, S.P.; Lobo, R.S.; Castilho, J.G.; Macedo, C.I.; Carnieli, P., Jr.; Fahl, W.O.; Achkar, S.M.; Scheffer, W.; Kotait, I.; et al. Rabies Virus in Insectivorous Bats: Implications of the Diversity of the Nucleoprotein and Glycoprotein Genes for Molecular Epidemiology. Virology 2010, 405, 352–360. [Google Scholar] [CrossRef]
- Yang, Y.J.; Zhao, P.S.; Wu, H.X.; Wang, H.L.; Zhao, L.L.; Xue, X.H.; Gai, W.W.; Gao, Y.W.; Yang, S.T.; Xia, X.Z. Production and Characterization of a Fusion Peptide Derived from the Rabies Virus Glycoprotein (RVG29). Protein Expr. Purif. 2014, 104, 7–13. [Google Scholar] [CrossRef]
- Nitschel, S.; Zaeck, L.M.; Potratz, M.; Nolden, T.; Kamp, V.; Franzke, K.; Höper, D.; Pfaff, F.; Finke, S. Point Mutations in the Glycoprotein Ectodomain of Field Rabies Viruses Mediate Cell Culture Adaptation through Improved Virus Release in a Host Cell Dependent and Independent Manner. Viruses 2021, 13, 1989. [Google Scholar] [CrossRef]

| Passage | Tissue | SNPs (>50%) | Minor Variants (>20%) | ||||
|---|---|---|---|---|---|---|---|
| A2397G | A2413C | T2699C | T2977Y | T3445G | |||
| OM | Medulla oblongata | ||||||
| Spinal cord | |||||||
| Hippocampus | |||||||
| P1 | Medulla oblongata | ||||||
| Spinal cord | |||||||
| Hippocampus | |||||||
| P3 | Medulla oblongata | ||||||
| Spinal cord | T2977Y (M2: S185P) | 21.6 | |||||
| Hippocampus | A2399G (non-coding) | ||||||
| P5 | Medulla oblongata | G2397A (non-coding) | 43.4 | 20.8 | 20.1 | ||
| Spinal cord | T2977C (M2: S185P) | >50 | |||||
| Hippocampus | A2399G (non-coding) | ||||||
| Mouse ID | Organ | SNPs (>50%) | Minor Variants (>20%) | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A247C | C2406A | T4600C (G:R456W) | A5315G (L:G32D) | T5898G (L:D226E) | A6771G | A7704G | G7704A | A8313G | T10668C | C10668T | G10785A | C11031T | T11031C | A11775G | G11775A | |||
| 2 | Pharyngeal swab | A1617C (M1:L62T), C10668T | ||||||||||||||||
| 3 | Salivary gland | C1783T | ||||||||||||||||
| 3 | Pharyngeal swab | C1702T, T5898G (L:D226E) | ||||||||||||||||
| 5 | Trigeminal ganglion | G3131T | ||||||||||||||||
| 5 | Spinal cord—dorsal | G3131T | ||||||||||||||||
| 5 | Spinal cord—ventral | G3131T | ||||||||||||||||
| 6 | Brain | C247A | ||||||||||||||||
| 6 | Trigeminal ganglion | C247A | ||||||||||||||||
| 6 | Spinal cord—dorsal | C247A | ||||||||||||||||
| 6 | Spinal cord—ventral | C247A | ||||||||||||||||
| 6 | Nuchal skin | C247A, C2552T | ||||||||||||||||
| 7 | Brain | A2406C, C5246T | ||||||||||||||||
| 7 | Trigeminal ganglion | A2406C, C5246T | ||||||||||||||||
| 7 | Spinal cord—dorsal | A2406C, C5246T | ||||||||||||||||
| 7 | Spinal cord—ventral | A2406C, C5246T | ||||||||||||||||
| 7 | Nuchal skin | A2406C | ||||||||||||||||
| 8 | Brain | |||||||||||||||||
| 8 | Trigeminal ganglion | C10668T | ||||||||||||||||
| 8 | Spinal cord—dorsal | |||||||||||||||||
| 8 | Spinal cord—ventral | |||||||||||||||||
| 8 | Nuchal skin | |||||||||||||||||
| 8 | Tongue epithelium | C10668T | ||||||||||||||||
| 8 | Salivary gland | |||||||||||||||||
| 9 | Brain | |||||||||||||||||
| 9 | Trigeminal ganglion | |||||||||||||||||
| 9 | Spinal cord—dorsal | |||||||||||||||||
| 9 | Spinal cord—ventral | |||||||||||||||||
| 9 | Nuchal skin | T4600C (G:R456W) | ||||||||||||||||
| 9 | Tongue epithelium | |||||||||||||||||
| 9 | Salivary gland | |||||||||||||||||
| 10 | Brain | |||||||||||||||||
| 10 | Trigeminal ganglion | |||||||||||||||||
| 10 | Spinal cord—dorsal | A7704G, C11031T, G11775A | ||||||||||||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Embregts, C.W.E.; Farag, E.A.B.A.; Bansal, D.; Boter, M.; van der Linden, A.; Vaes, V.P.; van Middelkoop-van den Berg, I.; IJpelaar, J.; Ziglam, H.; Coyle, P.V.; et al. Rabies Virus Populations in Humans and Mice Show Minor Inter-Host Variability within Various Central Nervous System Regions and Peripheral Tissues. Viruses 2022, 14, 2661. https://doi.org/10.3390/v14122661
Embregts CWE, Farag EABA, Bansal D, Boter M, van der Linden A, Vaes VP, van Middelkoop-van den Berg I, IJpelaar J, Ziglam H, Coyle PV, et al. Rabies Virus Populations in Humans and Mice Show Minor Inter-Host Variability within Various Central Nervous System Regions and Peripheral Tissues. Viruses. 2022; 14(12):2661. https://doi.org/10.3390/v14122661
Chicago/Turabian StyleEmbregts, Carmen W. E., Elmoubashar A. B. A. Farag, Devendra Bansal, Marjan Boter, Anne van der Linden, Vincent P. Vaes, Ingeborg van Middelkoop-van den Berg, Jeroen. IJpelaar, Hisham Ziglam, Peter V. Coyle, and et al. 2022. "Rabies Virus Populations in Humans and Mice Show Minor Inter-Host Variability within Various Central Nervous System Regions and Peripheral Tissues" Viruses 14, no. 12: 2661. https://doi.org/10.3390/v14122661








