Transmission Dynamics of Punique Virus in Tunisia
Abstract
1. Introduction
2. Material and Methods
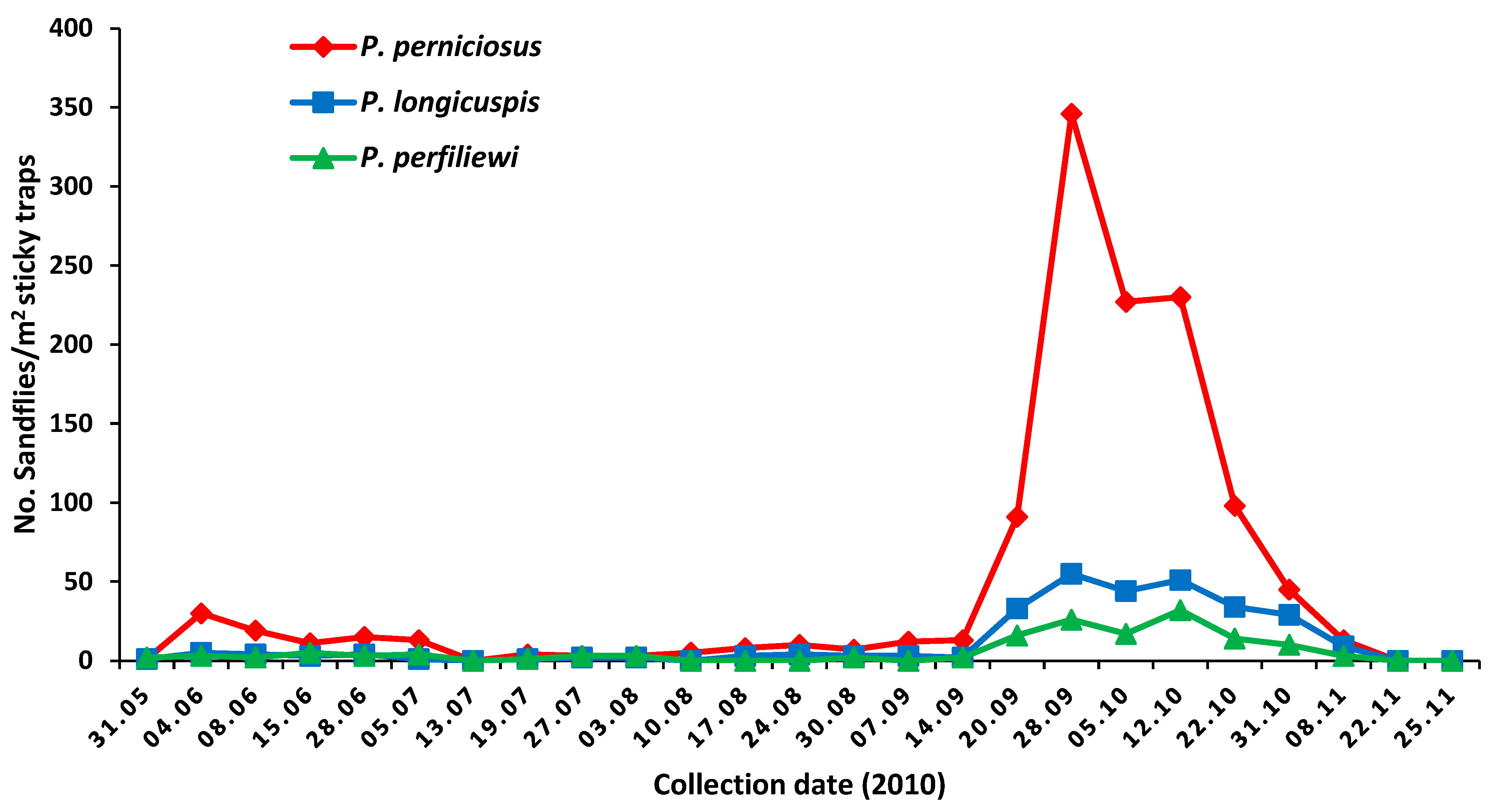
2.1. Collection of Sandflies
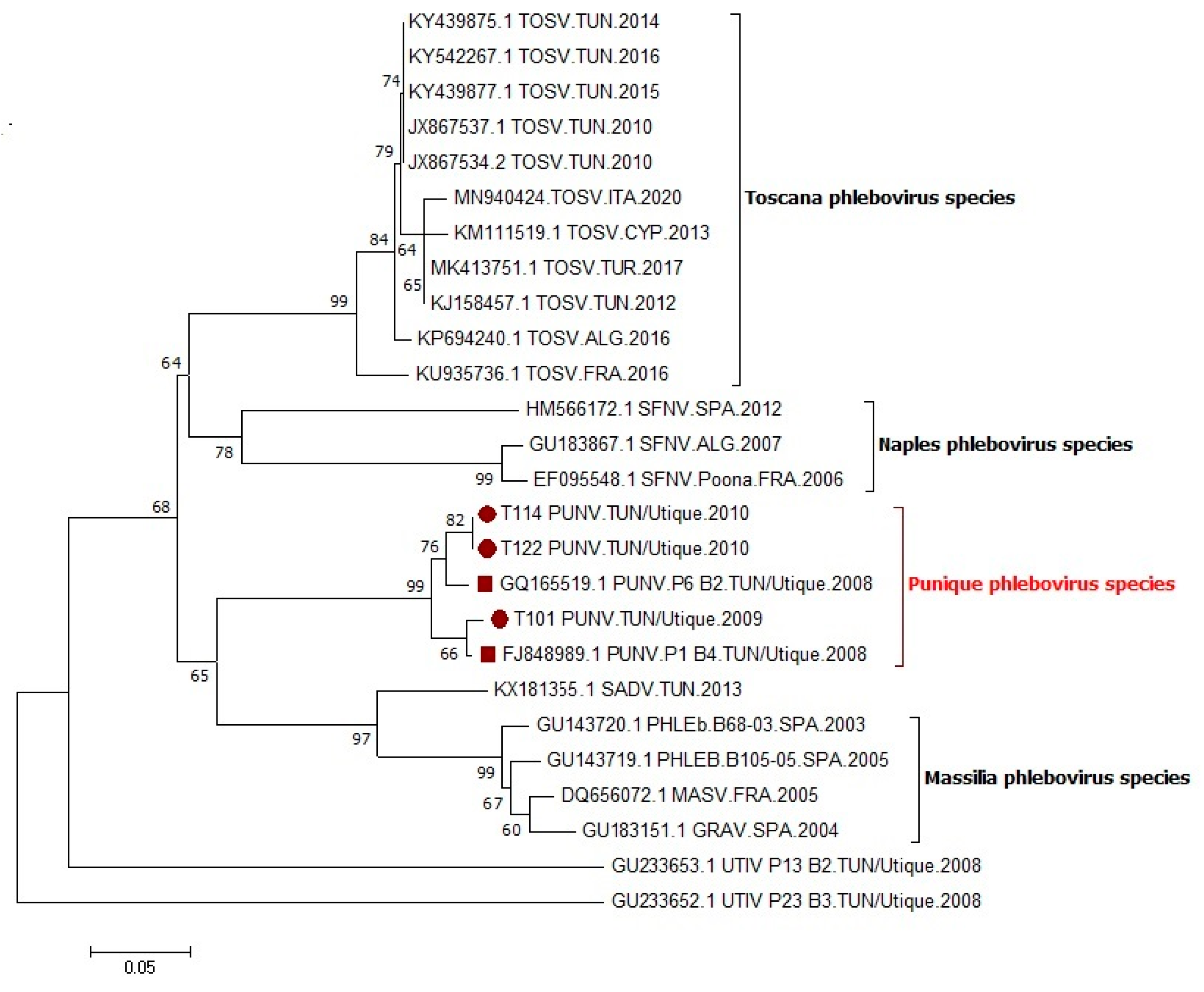
2.2. Virus Detection, Isolation, Sequencing, and Phylogenetic Analysis
3. Results
3.1. Sandfly Trapping and Virus Detection
3.2. Virus Isolation and Phylogenetic Study
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Alkan, C.; Bichaud, L.; de Lamballerie, X.; Alten, B.; Gould, E.A.; Charrel, R.N. Sandfly-borne phleboviruses of Eurasia and Africa: Epidemiology, genetic diversity, geographic range, control measures. Antivir. Res. 2013, 100, 54–74. [Google Scholar] [CrossRef] [PubMed]
- Charrel, R.N.; Gallian, P.; Navarro-Mari, J.M.; Nicoletti, L.; Papa, A.; Sanchez-Seco, M.P.; Tenorio, A.; de Lamballerie, X. Emergence of Toscana virus in Europe. Emerg. Infect. Dis. 2005, 11, 1657–1663. [Google Scholar] [CrossRef]
- Anagnostou, V.; Pardalos, G.; Athanasiou-Metaxa, M.; Papa, A. Novel phlebovirus in febrile child, Greece. Emerg. Infect. Dis. 2011, 17, 940–941. [Google Scholar] [CrossRef] [PubMed]
- Bahri, O.; Fazaa, O.; Ben Alaya-Bouafif, N.; Bouloy, M.; Triki, H.; Bouattour, A. Role of Toscana virus in meningo-encephalitis in Tunisia. Pathol. Biol. 2011, 59, 125–127. [Google Scholar]
- Ergunay, K.; Ismayilova, V.; Colpak, I.A.; Kansu, T.; Durdal Us, D. A case of central nervous system infection due to a novel Sandfly Fever Virus (SFV) variant: Sandfly Fever Turkey Virus (SFTV). J. Clin. Virol. 2012, 54, 79–82. [Google Scholar] [CrossRef]
- Nougairede, A.; Bichaud, L.; Thiberville, S.D.; Ninove, L.; Zandotti, C.; de Lamballerie, X.; Brouqui, P.; Charrel, R.N. Isolation of Toscana virus from the cerebrospinal 1 fluid of a man with meningitis in Marseille, France, 2010. Vector Borne Zoonotic. Dis. 2013, 13, 685–688. [Google Scholar] [CrossRef]
- Depaquit, J.; Grandadam, M.; Fouque, F.; Andry, P.E.; Peyrefitte, C. Arthropod- borne viruses transmitted by Phlebotominae sandflies in Europe: A review. Eur. Surveill. 2010, 15, 19507. [Google Scholar] [CrossRef]
- Ergunay, K.; Erisoz Kasap, O.; Orsten, S.; Oter, K.; Gunay, F.; Akkutay, A.Z.; Dincer, E.; Alten, B.; Ozkul, A. Phlebovirus and Leishmania detection in sandflies from eastern Thrace and Northern Cyprus. Parasite Vectors 2014, 12, 575. [Google Scholar] [CrossRef]
- Charrel, R.N.; Moureau, G.; Temmam, S.; Izri, A.; Marty, P.; Parola, P.; da Rosa, A.T.; Tesh, R.B.; de Lamballerie, X. Massilia virus, a novel Phlebovirus (Bunyaviridae) isolated from sandflies in the Mediterranean. Vector-Borne Zoonotic Dis. 2009, 9, 519–530. [Google Scholar] [CrossRef]
- Amaro, F.; Zé-Zé, L.; Lourenço, L.; Giovanetti, M.; Becker, S.C.; Alves, M.J. Phylogenetic analysis of Massilia phlebovirus in Portugal. Viruses 2021, 13, 1412. [Google Scholar] [CrossRef]
- Collao, X.; Palacios, G.; de Ory, F.; Sanbonmatsu, S.; Perez-Ruiz, M.; Navarro, J.M.; Molina, R.; Hutchison, S.K.; Lipkin, W.I.; Tenorio, A.; et al. Granada virus: A natural phlebovirus reassortant of the sandfly fever Naples serocomplex with low seroprevalence in humans. Am. J. Trop. Med. Hyg. 2010, 83, 760–765. [Google Scholar] [CrossRef]
- Papa, A.; Velo, E.; Bino, S. A novel phlebovirus in Albanian sandflies. Clin. Microbiol. Infect. 2011, 17, 585–587. [Google Scholar] [CrossRef] [PubMed]
- Peyrefitte, C.N.; Grandadam, M.; Bessaud, M.; Andry, P.E.; Fouque, F.; Caro, V.; Diancourt, L.; Schuffenecker, I.; Pagès, F.; Tolou, H.; et al. Diversity of Phlebotomus perniciosus in Provence, Southeastern France: Detection of Two Putative New Phlebovirus Sequences. Vector-Borne Zoonotic. Dis. 2013, 13, 630–636. [Google Scholar] [CrossRef]
- Remoli, M.E.; Fortuna, C.; Marchi, A.; Bucci, P.; Argentini, C.; Bongiorno, G.; Maroli, M.; Gradoni, L.; Gramiccia, M.; Ciufolini, M.G. Viral isolates of a novel putative phlebovirus in the Marche Region of Italy. Am. J. Trop. Med. Hyg. 2014, 90, 760–763. [Google Scholar] [CrossRef]
- Izri, A.; Temmam, S.; Moureau, G.; Hamrioui, B.; de Lamballerie, X.; Charrel, R.N. Sandfly fever Sicilian virus, Algeria. Emerg. Infect. Dis. 2008, 14, 795–797. [Google Scholar] [CrossRef] [PubMed]
- Moureau, G.; Bichaud, L.; Salez, N.; Ninove, L.; Hamrioui, B.; Belazzoug, S.; de Lamballerie, X.; Izri, A.; Charrel, R.N. Molecular and serological evidence for the presence of novel phleboviruses in sandflies from northern Algeria. Open Virol. J. 2010, 4, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Es-Sette, N.; Nourlil, J.; Hamdi, S.; Mellouki, F.; Lemrani, M. First detection of Toscana virus RNA from sand flies in the genus Phlebotomus (Diptera: Phlebotomidae) naturally infected in Morocco. J. Med. Entomol. 2012, 49, 1507–1509. [Google Scholar] [CrossRef] [PubMed]
- Alkan, C.; Allal-Ikhlef, A.L.; Alwassouf, S.; Baklouti, A.; Piorkowski, G.; de Lamballerie, X.; Charrel, R.N. Virus isolation, genetic characterization, and seroprevalence of Toscana virus in Algeria. Clin. Microbiol. Infect. 2015, 21, 1040. [Google Scholar] [CrossRef] [PubMed]
- Bichaud, L.; Dachraoui, K.; Piorkowski, G.; Chelbi, I.; Moureau, G.; Cherni, S.; de Lamballerie, X.; Sakhria, S.; Charrel, R.N.; Zhioua, E. 2013. Isolation of Toscana virus from sand flies, Tunisia. Emerg. Infect. Dis. 2013, 19, 322–324. [Google Scholar] [CrossRef] [PubMed]
- Dachraoui, K.; Fares, W.; Bichaud, L.; Barhoumi, W.; Beier, J.C.; Derbali, M.; Cherni, S.; de Lamballerie, X.; Chelbi, I.; Charrel, R.N.; et al. Phleboviruses associated with sand flies in arid bio-geographical areas of Central Tunisia. Acta Trop. 2016, 158, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Zhioua, E.; Moureau, G.; Chelbi, I.; Ninove, L.; Salez, N.; Derbali, M.; Champs, M.; Cherni, S.; Lamballerie, X.; Charrel, R.N. Punique virus, a novel phlebovirus, related to Sandfly fever Naples virus, isolated from sandflies collected in Tunisia. J. Gen. Virol. 2010, 91, 1275–1283. [Google Scholar] [CrossRef]
- Bichaud, L.; Dachraoui, K.; Alwassouf, S.; Alkan, C.; Mensi, M.; Piorkowski, G.; Sakhria, S.; Seston, M.; Fares, W.; de Lamballerie, X.; et al. Isolation, full genomic characterisation and neutralisation-based human seroprevalence of Medjerda Valley virus, a novel sandfly-borne phlebovirus belonging to the Salehabad virus complex in northern Tunisia. J. Gen. Virol. 2016, 97, 602–610. [Google Scholar] [CrossRef]
- Fares, W.; Charrel, R.N.; Dachraoui, K.; Bichaud, L.; Barhoumi, W.; Derbali, M.; Cherni, S.; Chelbi, I.; de Lamballerie, X.; Zhioua, E. Infection of sand flies collected from different bio-geographical areas of Tunisia with phleboviruses. Acta Trop. 2015, 141, 1–6. [Google Scholar] [CrossRef]
- Zhioua, E.; Kaabi, B.; Chelbi, I. Entomological investigations following the spread of visceral leishmaniasis toward the center and the south of Tunisia. J. Vector Ecol. 2007, 32, 371–374. [Google Scholar] [CrossRef]
- Chelbi, I.; Derbali, M.; Al-Ahmadi, Z.; Zaafouri, B.; El Fahem, A.; Zhioua, E. Phenology of Phlebotomus papatasi (Diptera: Psychodidae) relative to the seasonal prevalence of zoonotic cutaneous leishmaniasis in central Tunisia. J. Med. Entomol. 2007, 44, 385–388. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Croset, H.; Rioux, J.A.; Master, M.; Bayar, N. Les phlébotomes de la Tunisie (Diptera, Phlebotominae). Mise au point systématique, chorologique et éthologique. Ann. Parasitol. Hum. Comp. 1978, 53, 711–749. [Google Scholar] [CrossRef] [PubMed]
- Pesson, B.; Ready, J.S.; Benabdennbi, I.; Martín-Sánchez, J.; Esseghir, S.; Cadi-Soussi, M.; Morillas-Marquez, F.; Ready, P.D. Sandflies of the Phlebotomus perniciosus complex: Mitochondrial introgression and a new sibling species of Phlebotomus longicuspis in the Moroccan Rif. Med. Vet. Entomol. 2004, 18, 25–37. [Google Scholar] [CrossRef]
- Boussaa, S.; Boumezzough, A.; Remy, P.E.; Glasser, N.; Pesson, B. Morphological and isoenzymatic differentitation of Phlebotomus pernicious and Phlebotomus longicuspis (Diptera: Psychodidae) in Southern Morocco. Acta Trop. 2008, 106, 184–189. [Google Scholar] [CrossRef]
- Sanchez-Seco, M.P.; Echevarria, J.M.; Hernandez, L.; Estevez, D.; Navarro-Mari, J.M.; Tenorio, A. Detection and identification of Toscana and other phleboviruses by RT-nested-PCR assays 453 with degenerated primers. J. Med. Virol. 2003, 71, 140–149. [Google Scholar] [CrossRef] [PubMed]
- Chelbi, I.; Maghraoui, K.; Zhioua, S.; Cherni, S.; Labidi, I.; Satoskar, A.; Hamilton, J.G.C.; Zhioua, E. Enhanced attractiveness of sand fly vectors of Leishmania infantum to dogs infected with zoonotic visceral leishmaniasis. PLoS Negl. Trop. Dis. 2021, 15, e0009647. [Google Scholar]
- Verani, P.; Ciufolini, M.G.; Caciolli, S.; Renzi, A.; Nicoletti, L.; Sabatinelli, G.; Bartolozzi, D.; Volpi, G.; Amaducci, L.; Coluzzi, M.; et al. Ecology of viruses isolated from sand flies in Italy and characterized of a new Phlebovirus (Arabia virus). Am. J. Trop. Med. Hyg. 1988, 38, 433–443. [Google Scholar] [CrossRef] [PubMed]
- Sanbonmatsu-Gamez, S.; Perez-Ruiz, M.; Collao, X.; Sanchez-Seco, M.P.; Morillas-Marquez, F.; de La Rosa-Fraile, M.; Navarro-Mari, J.M.; Tenorio, A. Toscana virus in Spain. Emerg. Infect. Dis. 2005, 11, 1701–1707. [Google Scholar] [CrossRef] [PubMed]
- Charrel, R.N.; Izri, A.; Temmam, S.; Delaunay, P.; Toga, I.; Dumon, H.; Marty, P.; de Lamballerie, X.; Parolla, P. Cocirculation of 2 genotypes of Toscana virus, southeastern France. Emerg. Infect. Dis. 2007, 13, 465–468. [Google Scholar] [CrossRef] [PubMed]
- Calisher, C.H.; Calzolari, M. Taxonomy of phleboviruses, emphazising those that are sandfly-borne. Viruses 2021, 13, 918. [Google Scholar] [CrossRef] [PubMed]
- Tesh, R.B.; Modi, G.B. Maintenance of Toscana virus in Phlebotomus perniciosus by vertical transmission. Am. J. Trop. Med. Hyg. 1987, 36, 189–193. [Google Scholar] [CrossRef] [PubMed]
- Ciufolini, M.G.; Maroli, M.; Verani, P. Growth of two phleboviruses after experimental infection of their suspected sand fly vector, Phlebotomus perniciosus (Diptera: Psychodidae). Am. J. Trop. Med. Hyg. 1985, 34, 174–179. [Google Scholar] [CrossRef]
- Ciufolini, M.G.; Maroli, M.; Guandalini, E.; Marchi, A.; Verani, P. Experimental studies on the maintenance of Toscana and Arbia viruses (Bunyaviridae: Phlebovirus). Am. J. Trop. Med. Hyg. 1989, 40, 669–675. [Google Scholar] [CrossRef] [PubMed]
- Ciufolini, M.G.; Maroli, M.; Verani, P. Laboratory reared sandflies (Diptera: Psychodidae) and studies on phleboviruses. Parassitologia 1991, 33, 137–142. [Google Scholar] [PubMed]
- Maroli, M.; Ciufolini, M.G.; Verani, P. Vertical transmission of Toscana virus in the sandfly, Phlebotomus perniciosus, via the second gonotrophic cycle. Med. Vet. Entomol. 1993, 7, 283–286. [Google Scholar] [CrossRef] [PubMed]
- Jancarova, M.; Bichaud, L.; Hlavacova, J.; Priet, S.; Ayhan, N.; Spitzova, T.; Volf, P.; Charrel, R.N. Experimental infection of sand flies by Massilia virus and viral transmission by co-feeding on sugar meal. Viruses 2019, 11, 332. [Google Scholar] [CrossRef] [PubMed]
- Dolmatova, A.V.; Demina, N.A. Les Phlébotomes (Phlebotominae) et les Maladies qu’ils Transmettent; Office de la Recherche Scientifique et Technique d’Outre Mer (ORSTOM): Paris, France, 1971; 168p. [Google Scholar]
- Chelbi, I.; Zhioua, E. Biologiy of Phlebotomus papatasi (Diptera: Psychodidae) in the laboratory. J. Med. Entomol. 2007, 44, 597–600. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Maroli, M.; Fiorentino, S.; Guandalini, E. Biology of a laboratory colony of Phlebotomus perniciosus (Diptera: Psychodidae). J. Med. Entomol. 1987, 24, 547–551. [Google Scholar] [CrossRef]
- Muñoz, C.; Ayhan, N.; Ortuño, M.; Ortiz, J.; Gould, E.A.; Maia, C.; Berriatua, E.; Charrel, R.N. Experimental Infection of Dogs with Toscana Virus and Sandfly Fever Sicilian Virus to Determine Their Potential as Possible Vertebrate Hosts. Microorganisms 2020, 8, 596. [Google Scholar] [CrossRef] [PubMed]
- Verani, P.; Ciufolini, M.G.; Nicoletti, L.; Balducci, M.; Sabatinelli, G.; Coluzzi, M.; Paci, P.; Amaducciet, L. Ecological and epidemiological studies of Toscana virus, an arbovirus isolated from Phlebotomus. Ann. Ist. Super. Sanità 1982, 18, 397–399. [Google Scholar] [PubMed]
- Sakhria, S.; Alwassouf, S.; Fares, W.; Bichaud, L.; Dachraoui, K.; Alkan, C.; Zoghlami, Z.; de Lamballerie, X.; Zhioua, E.; Charrel, R.N. Presence of sand-fly phlebovirus of two antigenic complexes (Sandfly fever Naples virus and Sandfly fever Sicilian virus) in two different bio-geographical regions of Tunisia demonstrated by a microneutralization-based sero-prevalence study in dogs. Parasite Vectors 2014, 7, 476. [Google Scholar]
- Sakhria, S.; Bichaud, L.; Mensi, M.; Salez, N.; Dachraoui, K.; Thirion, L.; Cherni, S.; Chelbi, I.; de Lamballerie, X.; Zhioua, E.; et al. Co-Circulation of Toscana Virus and Punique Virus in Northern Tunisia: A microneutralisation-based seroprevalence study. PLoS Negl. Trop. Dis. 2013, 7, e2429. [Google Scholar] [CrossRef] [PubMed]
- Ben Slimane, T.; Chouihi, E.; Ben Hadj Ahmed, S.; Chelbi, I.; Barhoumi, W.; Cherni, S.; Zoghlami, Z.; Gharbi, M.; Zhioua, E. An investigation on vertical transmission of Leishmania infantum in experimentally infected dogs and assessment of offspring’s infectiousness potential by xenodiagnosis. Vet. Parasitol. 2014, 15, 282–286. [Google Scholar] [CrossRef] [PubMed]
- Zoghlami, Z.; Chouihi, E.; Barhoumi, W.; Dachraoui, K.; Massoudi, N.; Ben Hele, K.; Habboul, Z.; Hadhri, M.H.; Limam, S.; Mhadhbi, M.; et al. Interactions between canine and human visceral leishmaniases in a holoendemic focus of Central Tunisia. Acta Trop. 2014, 139, 32–38. [Google Scholar] [CrossRef] [PubMed]
- Fares, W.; Dachraoui, K.; Barhoumi, W.; Cherni, S.; Chelbi, I.; Zhioua, E. Co-circulation of Toscana virus and Leishmania infantum in a focus of zoonotic visceral leishmaniasis from Central Tunisia. Acta Trop. 2020, 204, 105342. [Google Scholar] [CrossRef] [PubMed]
| Species | Sandflies Number | Total (%) | |
|---|---|---|---|
| Male | Female | ||
| P. perniciosus | 337 | 413 | 750 (85.91) |
| P. longicuspis | 63 | 37 | 100 (11.45) |
| P. perfiliewi | 5 | 5 | 10 (1.14) |
| P. papatasi | 4 | 6 | 10 (1.14) |
| S. minuta parotti | 3 | 0 | 3 (0.34) |
| Total | 412 | 461 | 873 (100) |
| Date of Collection | Species | Numbers of Sandflies | ||
|---|---|---|---|---|
| Male | Female | |||
| 25 May 2009 | P. perniciosus | 90 | 17 | (91.45) |
| P. longicuspis | 5 | 1 | (5.12) | |
| S. minuta parotti | 3 | 0 | (2.56) | |
| P. papatasi | 1 | 0 | (0.85) | |
| 26 June 2009 | P. perniciosus | 20 | 15 | (39.32) |
| P. longicuspis | 36 | 17 | (59.55) | |
| P. papatasi | 0 | 1 | (1.12) | |
| 21 July 2009 | P. perniciosus | 13 | 30 | (71.66) |
| P. longicuspis | 9 | 2 | (18.33) | |
| P. perfiliewi | 2 | 1 | (5) | |
| P. papatasi | 1 | 2 | (5) | |
| 23 July 2009 | P. perniciosus | 30 | 30 | (76.92) |
| P. longicuspis | 7 | 5 | (15.38) | |
| P. papatasi | 2 | 3 | (6.41) | |
| P. perfiliewi | 1 | 0 | (1.28) | |
| 25 September 2009 | P. perniciosus | 90 | 150 | (96.38) |
| P. longicuspis | 2 | 5 | (2.81) | |
| P. perfiliewi | 0 | 2 | (0.80) | |
| 7 October 2009 | P. perniciosus | 94 | 171 * | (94.64) |
| P. longicuspis | 4 | 7 | (3.92) | |
| P. perfiliewi | 2 | 2 | (1.42) | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dachraoui, K.; Chelbi, I.; Ben Said, M.; Ben Osman, R.; Cherni, S.; Charrel, R.; Zhioua, E. Transmission Dynamics of Punique Virus in Tunisia. Viruses 2022, 14, 904. https://doi.org/10.3390/v14050904
Dachraoui K, Chelbi I, Ben Said M, Ben Osman R, Cherni S, Charrel R, Zhioua E. Transmission Dynamics of Punique Virus in Tunisia. Viruses. 2022; 14(5):904. https://doi.org/10.3390/v14050904
Chicago/Turabian StyleDachraoui, Khalil, Ifhem Chelbi, Mourad Ben Said, Raja Ben Osman, Saifedine Cherni, Rémi Charrel, and Elyes Zhioua. 2022. "Transmission Dynamics of Punique Virus in Tunisia" Viruses 14, no. 5: 904. https://doi.org/10.3390/v14050904
APA StyleDachraoui, K., Chelbi, I., Ben Said, M., Ben Osman, R., Cherni, S., Charrel, R., & Zhioua, E. (2022). Transmission Dynamics of Punique Virus in Tunisia. Viruses, 14(5), 904. https://doi.org/10.3390/v14050904








