Nano-Sized Chimeric Human Papillomavirus-16 L1 Virus-like Particles Displaying Mycobacterium tuberculosis Antigen Ag85B Enhance Ag85B-Specific Immune Responses in Female C57BL/c Mice
Abstract
:1. Introduction
2. Methods and Materials
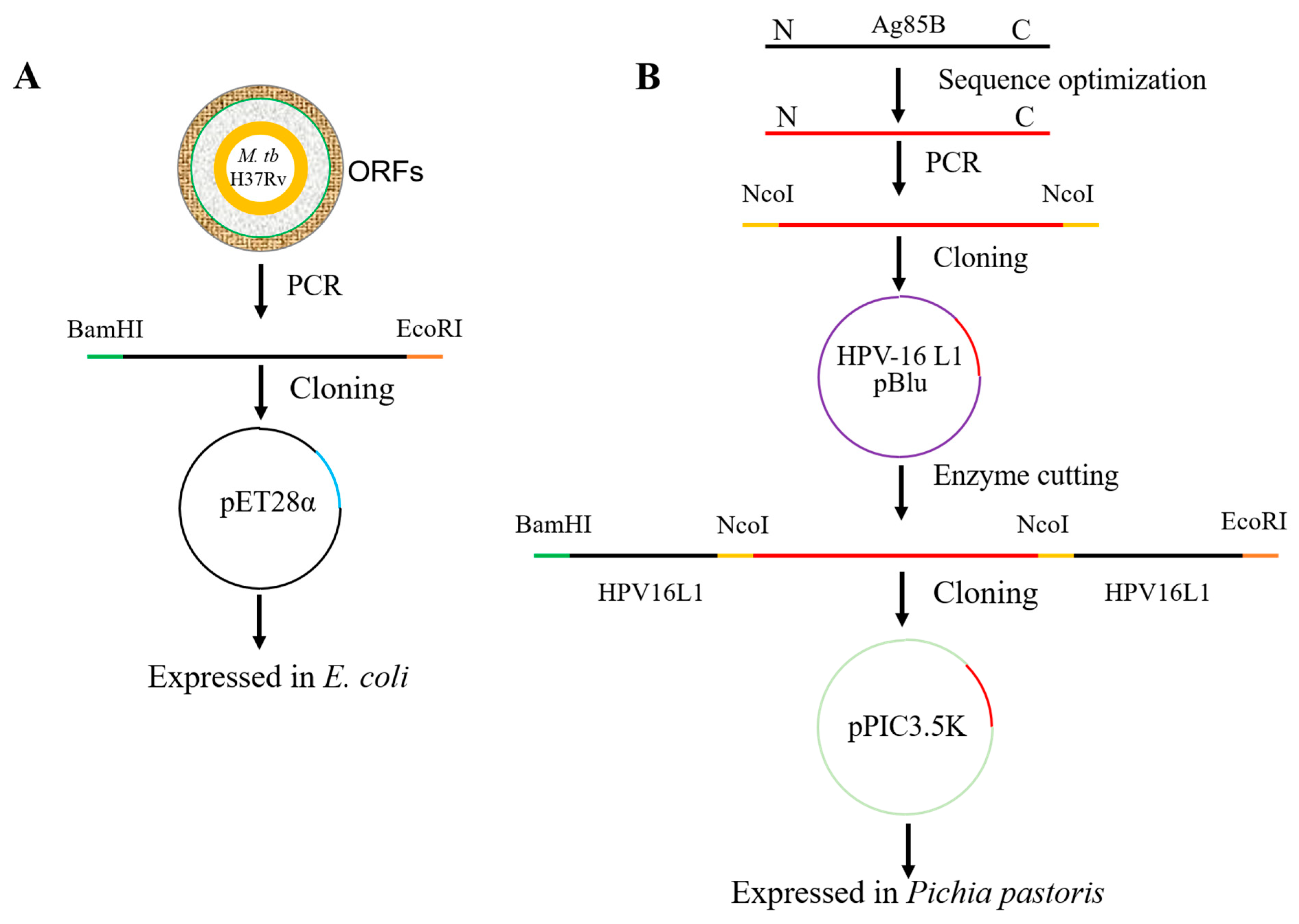
2.1. Cloning and Expression of Recombinant Ag85B in E. coli
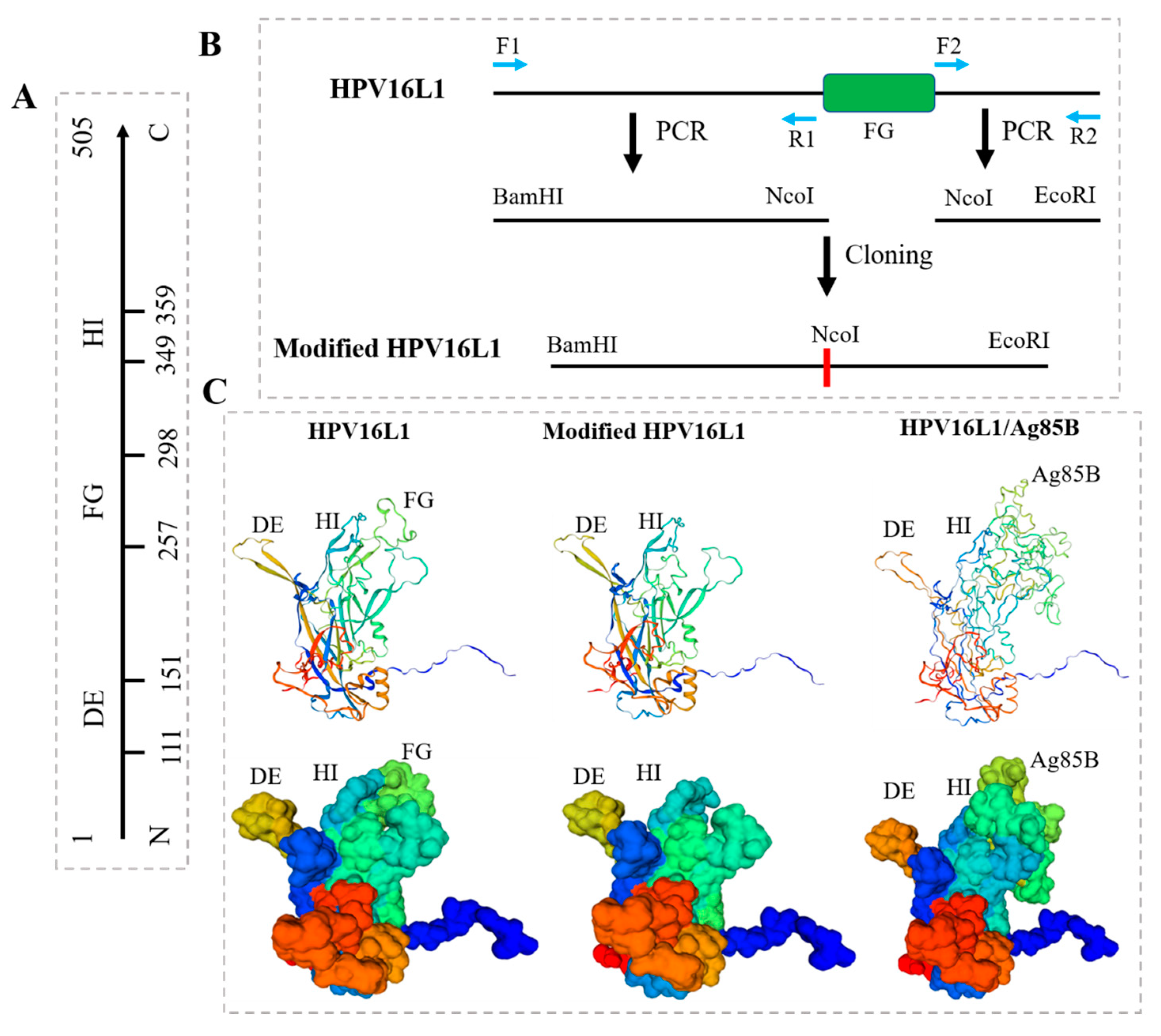
2.2. Modification of HPV16L1
2.3. Construction of Chimeric HPV16L1/Ag85B
2.4. Expression and Purification of HPV16L1/Ag85B VLP in Pichia Pastoris
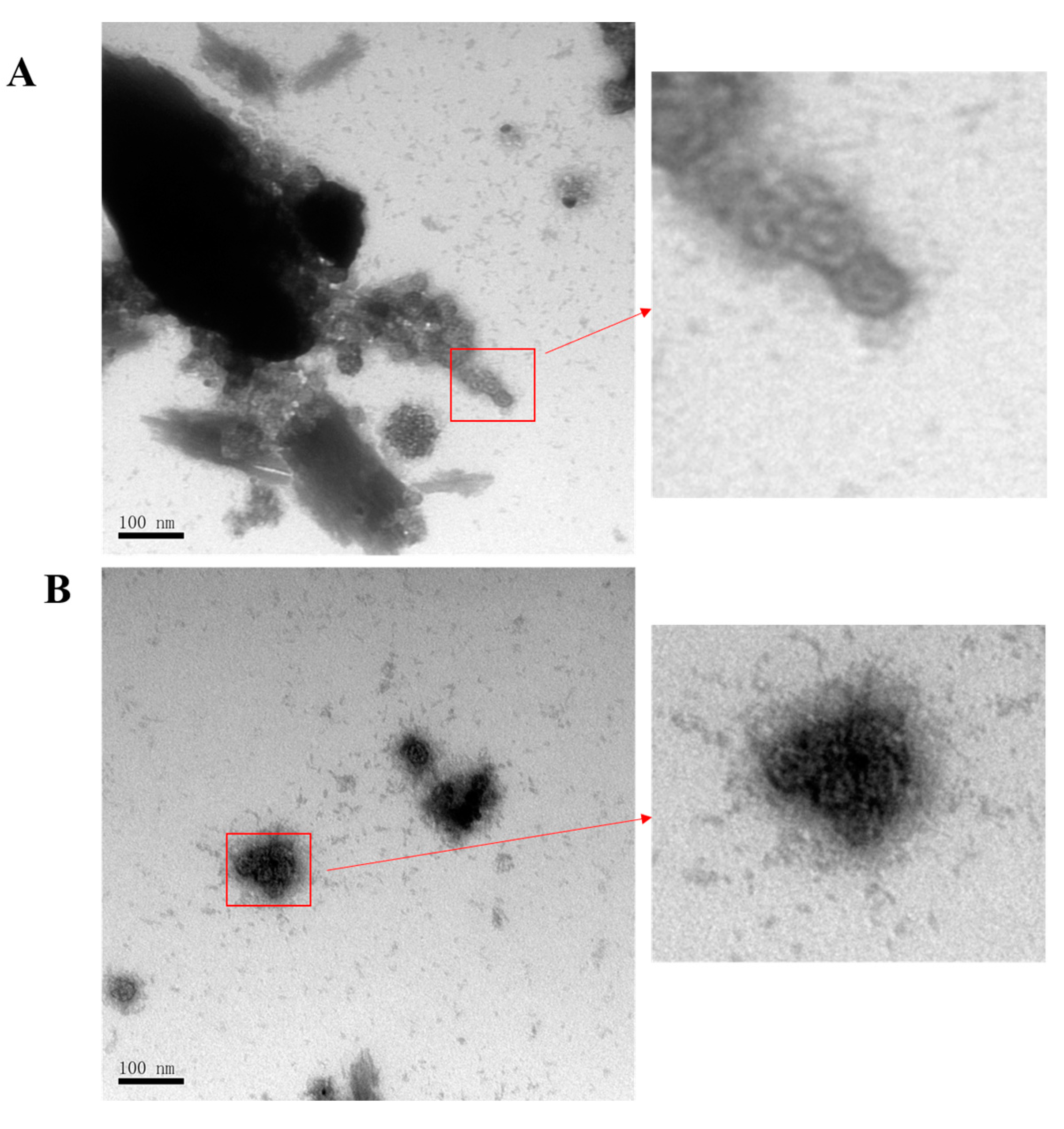
2.5. Negative Staining and Transmission Electron Microscopy (TEM)
2.6. Immunization of Mice, Collection of Sera, and Isolation of Splenocytes
2.7. Evaluation of Humoral Immune Response
2.8. Cytokine Analysis
2.9. Statistical Analysis
3. Results
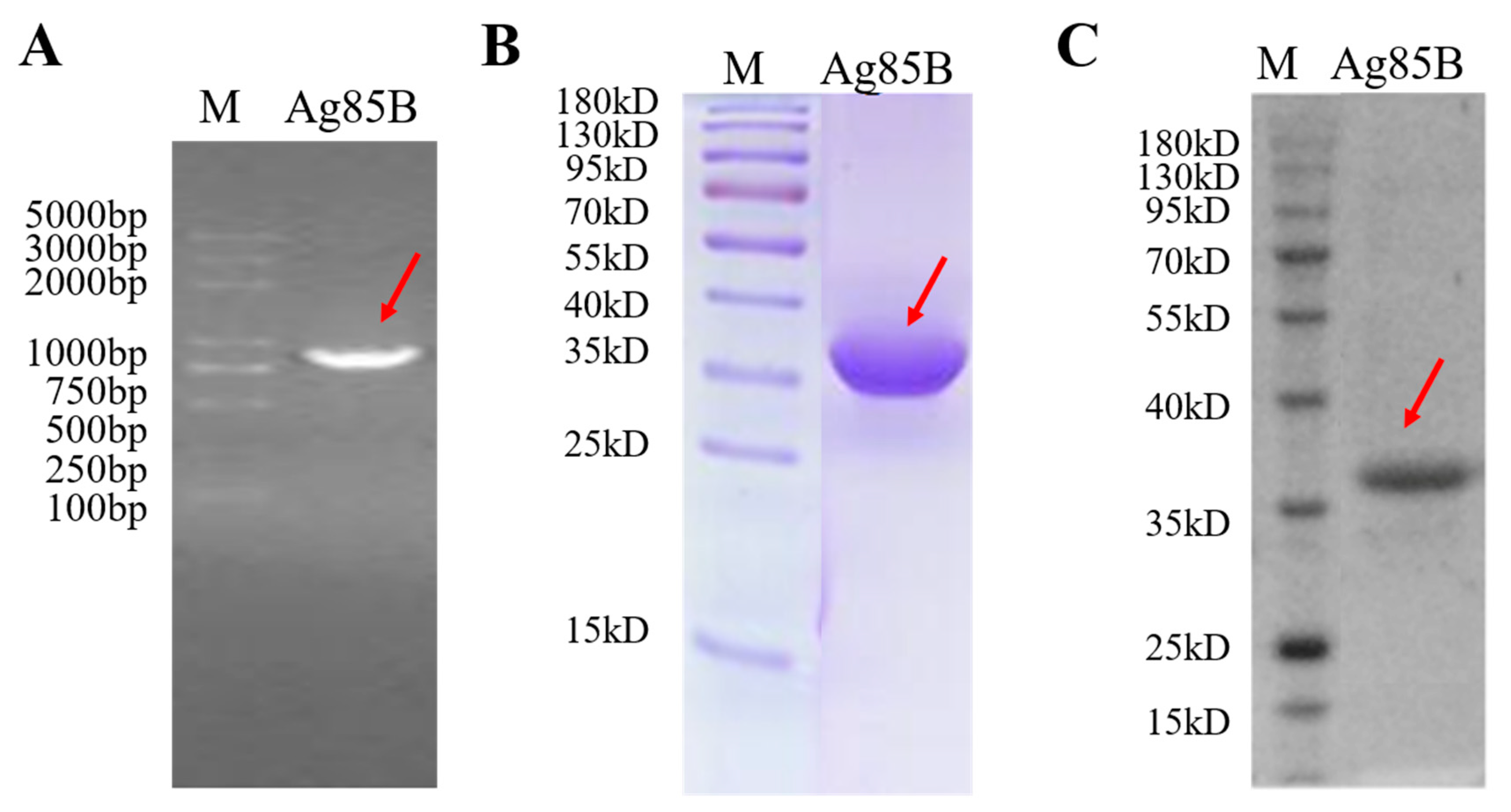
3.1. Characterization of Recombinant M. tb Antigen Ag85B
3.2. Modification of HPV16L1 and Construction of HPV16L1/Ag85B
3.3. Production of HPV16L1/Ag85B VLPs Using Pichia Pastoris Expression System
3.4. HPV16L1/Ag85B VLP Immunization Induced Antigen-Specific Antibody Responses
3.5. HPV16L1/Ag85B VLP Immunization Induced Significantly Higher Ag85B-Specific IFN-γ + T Cell Responses
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- World Health Organization. Global Tubeculosis Report 2022; World Health Organization: Geneva, Switzerland, 2022. [Google Scholar]
- McQuaid, C.F.; Vassall, A.; Cohen, T.; Fiekert, K.; White, R.G. The impact of COVID-19 on TB: A review of the data. Int. J. Tuberc. Lung Dis. 2021, 25, 436–446. [Google Scholar] [CrossRef]
- Venkatesan, P. Progress in tuberculosis vaccine research. Lancet Microbe 2021, 2, e12. [Google Scholar] [CrossRef]
- Mangtani, P.; Abubakar, I.; Ariti, C.; Beynon, R.; Pimpin, L.; Fine, P.E.; Rodrigues, L.C.; Smith, P.G.; Lipman, M.; Whiting, P.F.; et al. Protection by BCG vaccine against tuberculosis: A systematic review of randomized controlled trials. Clin. Infect. Dis. 2014, 58, 470–480. [Google Scholar] [CrossRef]
- Mohsen, K.Z.B.; Soleimanpour, S.; Rezaee, S.A. Antigen 85 complex as a powerful Mycobacterium tuberculosis immunogene: Biology, immune-pathogenicity, applications in diagnosis, and vaccine design. Microb. Pathog. 2017, 112, 20–29. [Google Scholar]
- Available online: https://newtbvaccines.org/tb-vaccine-pipeline/ (accessed on 1 October 2023).
- Lu, J.; Yang, L.; Su, C.; Shen, X.; Du, W. Therapeutic evaluation of antibiotics combined with recombinant tuberculosis vaccine AEC/BC02 in a guinea pig model of Mycobacterium tuberculosis infection. Chin. J. Microbiol. Immunol. 2018, 06, 414–419. [Google Scholar]
- Penn-Nicholson, A.; Tameris, M.; Smit, E.; Day, T.A.; Musvosvi, M.; Jayashankar, L.; Vergara, J.; Mabwe, S.; Bilek, N.; Geldenhuys, H.; et al. Safety and immunogenicity of the novel tuberculosis vaccine ID93 + GLA-SE in BCG-vaccinated healthy adults in South Africa: A randomised, double-blind, placebo-controlled phase 1 trial. Lancet Respir. Med. 2018, 6, 287–298. [Google Scholar] [CrossRef]
- Day, T.A.; Penn-Nicholson, A.; Luabeya, A.K.K.; Fiore-Gartland, A.; Du Plessis, N.; Loxton, A.G.; Vergara, J.; Rolf, T.A.; Reid, T.D.; Toefy, A.; et al. Safety and immunogenicity of the adjunct therapeutic vaccine ID93 + GLA-SE in adults who have completed treatment for tuberculosis: A randomised, double-blind, placebo-controlled, phase 2a trial. Lancet Respir. Med. 2021, 9, 373–386. [Google Scholar] [CrossRef] [PubMed]
- Suliman, S.; Luabeya, A.K.K.; Geldenhuys, H.; Tameris, M.; Hoff, S.T.; Shi, Z.; Tait, D.; Kromann, I.; Ruhwald, M.; Rutkowski, K.T.; et al. Dose Optimization of H56:IC31 Vaccine for Tuberculosis-Endemic Populations. A Double-Blind, Placebo-controlled, Dose-Selection Trial. Am. J. Respir. Crit. Care Med. 2019, 199, 220–231. [Google Scholar] [CrossRef] [PubMed]
- Van Der Meeren, O.; Hatherill, M.; Nduba, V.; Wilkinson, R.J.; Muyoyeta, M.; Van Brakel, E.; Ayles, H.M.; Henostroza, G.; Thienemann, F.; Scriba, T.J.; et al. Phase 2b Controlled Trial of M72/AS01E Vaccine to Prevent Tuberculosis. N Engl. J. Med. 2018, 379, 1621–1634. [Google Scholar] [CrossRef] [PubMed]
- Aagaard, C.; Hoang, T.; Dietrich, J.; Cardona, P.J.; Izzo, A.; Dolganov, G.; Schoolnik, G.K.; Cassidy, J.P.; Billeskov, R.; Andersen, P. A multistage tuberculosis vaccine that confers efficient protection before and after exposure. Nat. Med. 2011, 17, 189–194. [Google Scholar] [CrossRef] [PubMed]
- Tkachuk, A.P.; Gushchin, V.A.; Potapov, V.D.; Demidenko, A.V.; Lunin, V.G.; Gintsburg, A.L. Multi-subunit BCG booster vaccine GamTBvac: Assessment of immunogenicity and protective efficacy in murine and guinea pig TB models. PLoS ONE 2017, 12, e0176784. [Google Scholar] [CrossRef]
- Jeyanathan, M.; Damjanovic, D.; Yao, Y.; Bramson, J.; Smaill, F.; Xing, Z. Induction of an Immune-Protective T-Cell Repertoire With Diverse Genetic Coverage by a Novel Viral-Vectored Tuberculosis Vaccine in Humans. J. Infect. Dis. 2016, 214, 1996–2005. [Google Scholar] [CrossRef]
- Buzitskaya, Z.; Stosman, K.; Khairullin, B.; Kassenov, M.; Nurpeisova, A.; Sansyzbay, A.A.; Shurygina, A.P.; Aleksandrov, A.; Sivak, K.; Stukova, M. A New Intranasal Influenza Vector-Based Vaccine TB/FLU-04L Against Tuberculosis: Preclinical Safety Studies. Drug Res. 2022, 72, 255–258. [Google Scholar] [CrossRef]
- Minhinnick, A.; Satti, I.; Harris, S.; Wilkie, M.; Sheehan, S.; Stockdale, L.; Thomas, Z.R.M.; Lopez-Ramon, R.; Poulton, I.; Lawrie, A.; et al. A first-in-human phase 1 trial to evaluate the safety and immunogenicity of the candidate tuberculosis vaccine MVA85A-IMX313, administered to BCG-vaccinated adults. Vaccine 2016, 34, 1412–1421. [Google Scholar] [CrossRef]
- Li, W.; Li, M.; Deng, G.; Zhao, L.; Liu, X.; Wang, Y. Prime-boost vaccination with Bacillus Calmette Guerin and a recombinant adenovirus co-expressing CFP10, ESAT6, Ag85A and Ag85B of Mycobacterium tuberculosis induces robust antigen-specific immune responses in mice. Mol. Med. Rep. 2015, 12, 3073–3080. [Google Scholar] [CrossRef]
- Hu, Z.; Lu, S.H.; Lowrie, D.B.; Fan, X.Y. Research Advances for Virus-vectored Tuberculosis Vaccines and Latest Findings on Tuberculosis Vaccine Development. Front. Immunol. 2022, 13, 895020. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.; Bakhru, P.; Saikolappan, S.; Das, K.; Jagannath, C. An autophagy-inducing and TLR-2 activating BCG vaccine induces a robust protection against tuberculosis in mice. Npj Vaccines 2019, 4, 34. [Google Scholar] [CrossRef] [PubMed]
- Counoupas, C.; Pinto, R.; Nagalingam, G.; Britton, W.J.; Triccas, J.A. Protective efficacy of recombinant BCG over-expressing protective, stage-specific antigens of Mycobacterium tuberculosis. Vaccine 2018, 36, 2619–2629. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Wong, K.W.; Zhao, H.M.; Wen, H.L.; Ji, P.; Ma, H.; Wu, K.; Lu, S.H.; Li, F.; Li, Z.M.; et al. Sendai Virus Mucosal Vaccination Establishes Lung-Resident Memory CD8T Cell Immunity and Boosts BCG-Primed Protection against TB in Mice. Mol. Ther. J. Am. Soc. Gene Ther. 2017, 25, 1222–1233. [Google Scholar] [CrossRef] [PubMed]
- Nooraei, S.; Bahrulolum, H.; Hoseini, Z.S.; Katalani, C.; Hajizade, A.; Easton, A.J.; Ahmadian, G. Virus-like particles: Preparation, immunogenicity and their roles as nanovaccines and drug nanocarriers. J. Nanobiotechnology 2021, 19, 59. [Google Scholar] [CrossRef] [PubMed]
- Fuenmayor, J.; Godia, F.; Cervera, L. Production of virus-like particles for vaccines. N. Biotechnol. 2017, 39, 174–180. [Google Scholar] [CrossRef]
- Zhao, Q.; Li, S.; Yu, H.; Xia, N.; Modis, Y. Virus-like particle-based human vaccines: Quality assessment based on structural and functional properties. Trends Biotechnol. 2013, 31, 654–663. [Google Scholar] [CrossRef] [PubMed]
- Greenwood, B.; Doumbo, O.K. Implementation of the malaria candidate vaccine RTS, S/AS01. Lancet 2016, 387, 318–319. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Zhang, Y.; Lin, D.; Lin, D.; Zhang, J.; Xu, J.; Liu, Y.M.; Hu, F.; Qing, X.; Xia, C.; et al. Serodiagnosis of Schistosoma japonicum infection: Genome-wide identification of a protein marker, and assessment of its diagnostic validity in a field study in China. Lancet Infect. Dis. 2014, 14, 489–497. [Google Scholar] [CrossRef] [PubMed]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef] [PubMed]
- Kirnbauer, R.; Booy, F.; Cheng, N.; Lowy, D.R.; Schiller, J.T. Papillomavirus L1 major capsid protein self-assembles into virus-like particles that are highly immunogenic. Proc. Natl. Acad. Sci. USA 1992, 89, 12180–12184. [Google Scholar] [CrossRef]
- Chen, X.S.; Garcea, R.L.; Goldberg, I.; Casini, G.; Harrison, S.C. Structure of small virus-like particles assembled from the L1 protein of human papillomavirus 16. Mol. Cell 2000, 5, 557–567. [Google Scholar] [CrossRef]
- Ao, Z.; Chen, W.; Tan, J.; Cheng, Y.; Xu, Y.; Wang, L.; Yao, X. Lentivirus-Based Virus-Like Particles Mediate Delivery of Caspase 8 into Breast Cancer Cells and Inhibit Tumor Growth. Cancer Biother. Radiopharm. 2019, 34, 33–41. [Google Scholar] [CrossRef]
- Chen, C.W.; Saubi, N.; Joseph-Munne, J. Chimeric Human Papillomavirus-16 Virus-like Particles Presenting HIV-1 P18I10 Peptide: Expression, Purification, Bio-Physical Properties and Immunogenicity in BALB/c Mice. Int. J. Mol. Sci. 2023, 24, 8060. [Google Scholar] [CrossRef]
- Chen, C.W.; Saubi, N.; Kilpelainen, A.; Joseph-Munne, J. Chimeric Human Papillomavirus-16 Virus-like Particles Presenting P18I10 and T20 Peptides from HIV-1 Envelope Induce HPV16 and HIV-1-Specific Humoral and T Cell-Mediated Immunity in BALB/c Mice. Vaccines 2022, 11, 15. [Google Scholar] [CrossRef] [PubMed]
- Bissett, S.L.; Godi, A.; Beddows, S. The DE and FG loops of the HPV major capsid protein contribute to the epitopes of vaccine-induced cross-neutralising antibodies. Sci. Rep. 2016, 6, 39730. [Google Scholar] [CrossRef] [PubMed]
- Rodo, M.J.; Rozot, V.; Nemes, E.; Dintwe, O.; Hatherill, M.; Little, F.; Scriba, T.J. A comparison of antigen-specific T cell responses induced by six novel tuberculosis vaccine candidates. PLoS Pathog. 2019, 15, e1007643. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.L.; Chung, A.W.; Rosebrock, T.R.; Ghebremichael, M.; Yu, W.H.; Grace, P.S.; Schoen, M.K.; Tafesse, F.; Martin, C.; Leung, V.; et al. A Functional Role for Antibodies in Tuberculosis. Cell 2016, 167, 433–443.e414. [Google Scholar] [CrossRef]
- Li, H.; Wang, X.X.; Wang, B.; Fu, L.; Liu, G.; Lu, Y.; Cao, M.; Huang, H.; Javid, B. Latently and uninfected healthcare workers exposed to TB make protective antibodies against Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 2017, 114, 5023–5028. [Google Scholar] [CrossRef]
- Zhou, F.; Xu, X.; Cui, X.; Pan, W. Development and Evaluation of a Fusion Polyprotein Based on HspX and Other Antigen Sequences for the Serodiagnosis of Tuberculosis. Front. Immunol. 2021, 12, 726920. [Google Scholar] [CrossRef]
- Moguche, A.O.; Musvosvi, M.; Penn-Nicholson, A.; Plumlee, C.R.; Mearns, H.; Geldenhuys, H.; Smit, E.; Abrahams, D.; Rozot, V.; Dintwe, O.; et al. Antigen Availability Shapes T Cell Differentiation and Function during Tuberculosis. Cell Host Microbe 2017, 21, 695–706.e695. [Google Scholar] [CrossRef] [PubMed]
- Mohsen, M.O.; Bachmann, M.F. Virus-like particle vaccinology, from bench to bedside. Cell Mol. Immunol. 2022, 19, 993–1011. [Google Scholar] [CrossRef]
- Krammer, F.; Schinko, T.; Messner, P.; Palmberger, D.; Ferko, B.; Grabherr, R. Influenza virus-like particles as an antigen-carrier platform for the ESAT-6 epitope of Mycobacterium tuberculosis. J. Virol. Methods 2010, 167, 17–22. [Google Scholar] [CrossRef]
- Yin, Y.; Li, H.; Wu, S.; Dong, D.; Zhang, J.; Fu, L.; Xu, J.; Chen, W. Hepatitis B virus core particles displaying Mycobacterium tuberculosis antigen ESAT-6 enhance ESAT-6-specific immune responses. Vaccine 2011, 29, 5645–5651. [Google Scholar] [CrossRef]
- Dhanasooraj, D.; Kumar, R.A.; Mundayoor, S. Vaccine delivery system for tuberculosis based on nano-sized hepatitis B virus core protein particles. Int. J. Nanomed 2013, 8, 835–843. [Google Scholar]
- Dhanasooraj, D.; Kumar, R.A.; Mundayoor, S. Subunit Protein Vaccine Delivery System for Tuberculosis Based on Hepatitis B Virus Core VLP (HBc-VLP) Particles. Methods Mol. Biol. 2016, 1404, 377–392. [Google Scholar] [PubMed]



| Primer Name | Targeted Gene | Sequence |
|---|---|---|
| F1 | left side of FG loop | GGGGATCCAAAAAAATGTCTTTGTGGCTTCCATCCGAA GC |
| R1 | CCCCATGGGACAAACATCTGCTCACGTC | |
| F2 | right side of FG loop | CCCCATGGCCTACTCCATCCGGTAGTATG |
| R2 | GGGAATTCCTATTACAACTTTCTTTTC | |
| F3 | AOX1 | GACTGGTTCCAATTGACAAGC |
| R3 | GCAAATGGCATTCTGACATCC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, F.; Zhang, D. Nano-Sized Chimeric Human Papillomavirus-16 L1 Virus-like Particles Displaying Mycobacterium tuberculosis Antigen Ag85B Enhance Ag85B-Specific Immune Responses in Female C57BL/c Mice. Viruses 2023, 15, 2123. https://doi.org/10.3390/v15102123
Zhou F, Zhang D. Nano-Sized Chimeric Human Papillomavirus-16 L1 Virus-like Particles Displaying Mycobacterium tuberculosis Antigen Ag85B Enhance Ag85B-Specific Immune Responses in Female C57BL/c Mice. Viruses. 2023; 15(10):2123. https://doi.org/10.3390/v15102123
Chicago/Turabian StyleZhou, Fangbin, and Dongmei Zhang. 2023. "Nano-Sized Chimeric Human Papillomavirus-16 L1 Virus-like Particles Displaying Mycobacterium tuberculosis Antigen Ag85B Enhance Ag85B-Specific Immune Responses in Female C57BL/c Mice" Viruses 15, no. 10: 2123. https://doi.org/10.3390/v15102123





