Effects of US7 and UL56 on Cell-to-Cell Spread of Human Herpes Simplex Virus 1
Abstract
:1. Introduction
2. Materials and Methods
2.1. Viruses and Cell Lines
2.2. Plasmids and Antibodies
2.3. Sample Collection and Virus Isolation
2.4. Next Generation Sequencing (NGS)
2.5. Construction of Recombinant Virus
2.6. Recovery of Plaque Phenotype under Exogenous Expression
2.7. Syncytial Formation Assays
2.8. Bioinformatic Analysis
2.9. GD Copy Number Statistics in Genome Replication Stage
2.10. One-Step Growth Curve Analyses
2.11. Comparative Experiment on the Dynamics of Cell Invasion Stage
2.12. Relative mRNA Expression of US7 and UL56
3. Results
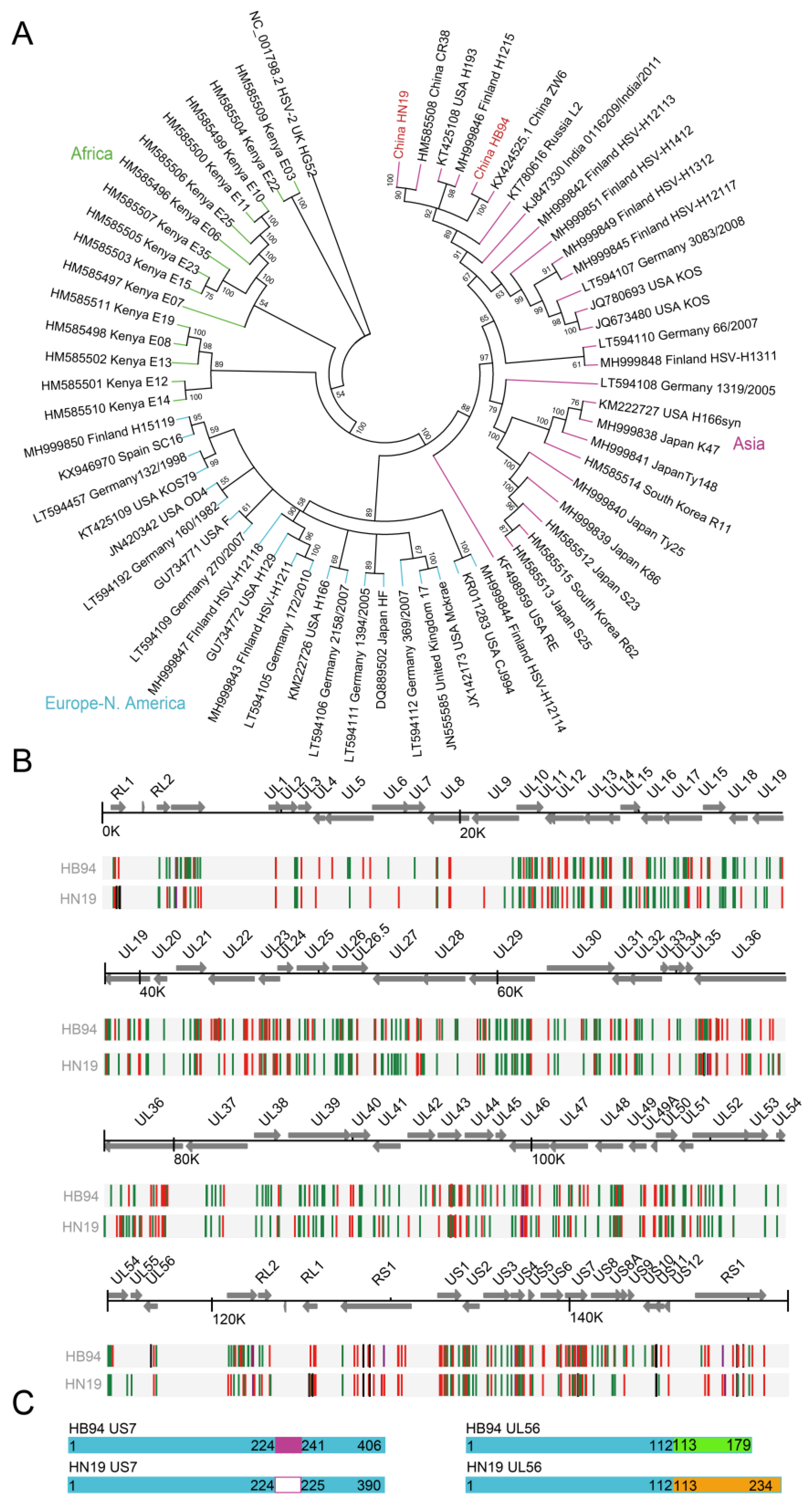
3.1. Stronger Cell-to-Cell Transmission Ability of HB94 Than that of HN19
3.2. US7 and UL56 Genes Showed Differences in HB94 and HN19 Strains via Phylogeny and Identity Analyses
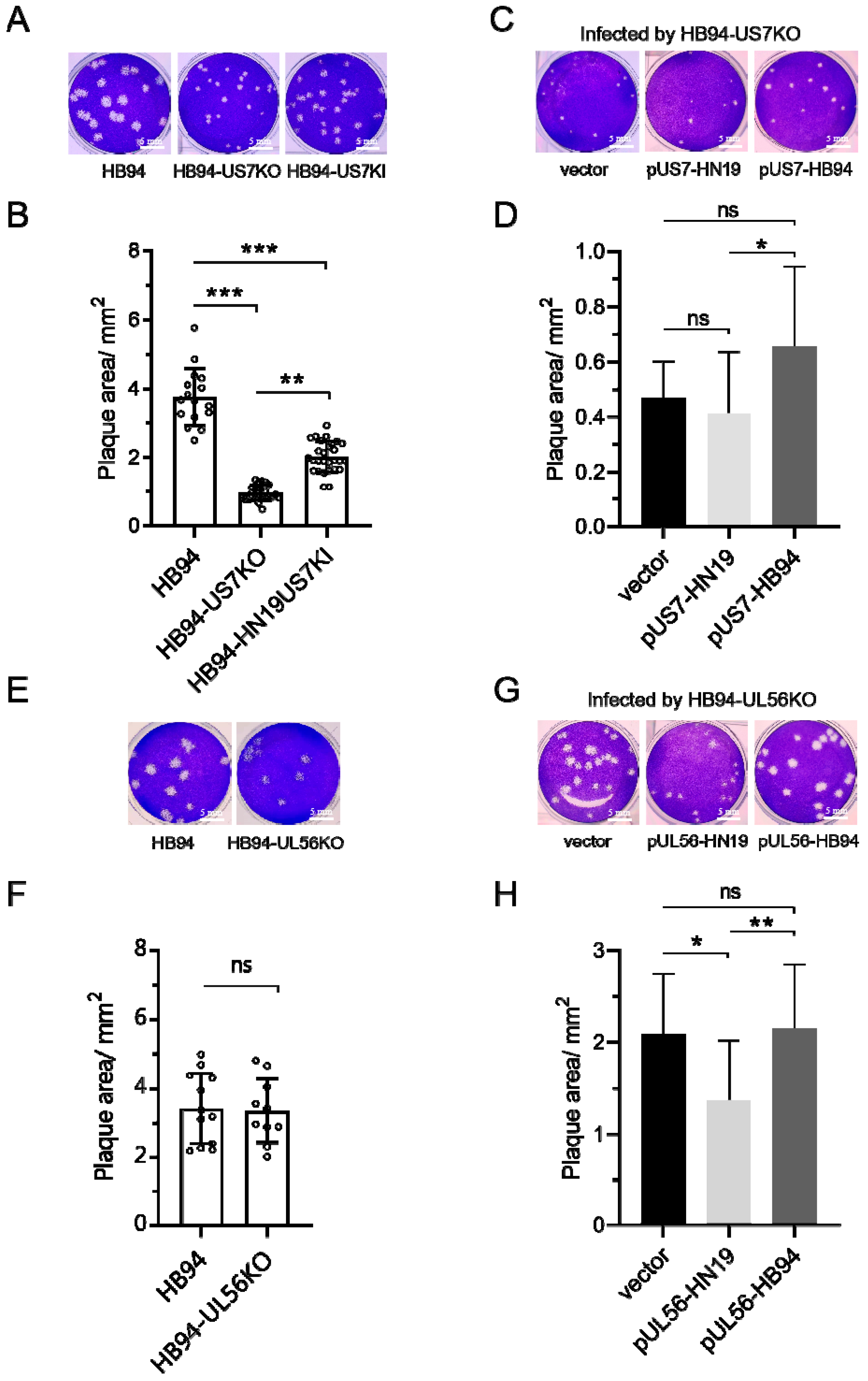
3.3. US7 Promoted the Formation of Plaques, and UL56 of HN19 Inhibited the Formation of Plaques
3.4. US7 Promoted Syncytial Formation, and UL56 Had no Effect on Syncytial Formation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Roizman, B.; Whitley, R.J. An Inquiry into the Molecular Basis of HSV Latency and Reactivation. Annu. Rev. Microbiol. 2013, 67, 355–374. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.S.; Robinson, N.J. Age-Specific Prevalence of Infection with Herpes Simplex Virus Types 2 and 1: A Global Review. J. Infect. Dis. 2002, 186, S3–S28. [Google Scholar] [CrossRef]
- Diefenbach, R.J.; Cornel, F. (Eds.) Herpes Simplex Virus: Methods and Protocols; Methods in Molecular Biology Series; Humana Press: New York, NY, USA, 2020. [Google Scholar]
- Chou, J.; Roizman, B. The terminal a sequence of the herpes simplex virus genome contains the promoter of a gene located in the repeat sequences of the L component. J. Virol. 1986, 57, 629–637. [Google Scholar] [CrossRef] [PubMed]
- Ventosa, M.; Ortiz-Temprano, A.; Khalique, H.; Lim, F. Synergistic effects of deleting multiple nonessential elements in nonreplicative HSV-1 BAC genomic vectors play a critical role in their viability. Gene Ther. 2017, 24, 433–440. [Google Scholar] [CrossRef] [PubMed]
- Velusamy, T.; Gowripalan, A.; Tscharke, D.C. CRISPR/Cas9-Based Genome Editing of HSV. Methods Mol. Biol. 2020, 2060, 169–183. [Google Scholar] [CrossRef] [PubMed]
- Manservigi, R.; Argnani, R.; Marconi, P. HSV Recombinant Vectors for Gene Therapy. Open Virol. J. 2010, 4, 123–156. [Google Scholar] [CrossRef]
- Ahmad, I.; Wilson, D.W. HSV-1 Cytoplasmic Envelopment and Egress. Int. J. Mol. Sci. 2020, 21, 5969. [Google Scholar] [CrossRef]
- Bowen, C.D.; Paavilainen, H.; Renner, D.W.; Palomäki, J.; Lehtinen, J.; Vuorinen, T.; Norberg, P.; Hukkanen, V.; Szpara, M.L. Comparison of Herpes Simplex Virus 1 Strains Circulating in Finland Demonstrates the Uncoupling of Whole-Genome Relatedness and Phenotypic Outcomes of Viral Infection. J. Virol. 2019, 93, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Szpara, M.L.; Gatherer, D.; Ochoa, A.; Greenbaum, B.; Dolan, A.; Bowden, R.J.; Enquist, L.W.; Legendre, M.; Davison, A.J. Evolution and diversity in human herpes simplex virus genomes. J. Virol. 2014, 88, 1209–1227. [Google Scholar] [CrossRef]
- Rice, S.A. Release of HSV-1 Cell-Free Virions: Mechanisms, Regulation, and Likely Role in Human-Human Transmission. Viruses 2021, 13, 2395. [Google Scholar] [CrossRef]
- Johnson, D.C.; Huber, M.T. Directed Egress of Animal Viruses Promotes Cell-to-Cell Spread. J. Virol. 2002, 76, 1–8. [Google Scholar] [CrossRef]
- Dingwell, K.S.; Brunetti, C.R.; Hendricks, R.L.; Tang, Q.; Tang, M.; Rainbow, A.J.; Johnson, D.C. Herpes simplex virus glycoproteins E and I facilitate cell-to-cell spread in vivo and across junctions of cultured cells. J. Virol. 1994, 68, 834–845. [Google Scholar] [CrossRef]
- Wang, F.; Zumbrun, E.E.; Huang, J.; Si, H.; Makaroun, L.; Friedman, H.M. Herpes simplex virus type 2 glycoprotein E is required for efficient virus spread from epithelial cells to neurons and for targeting viral proteins from the neuron cell body into axons. Virology 2010, 405, 269–279. [Google Scholar] [CrossRef] [PubMed]
- Carmichael, J.C.; Yokota, H.; Craven, R.C.; Schmitt, A.; Wills, J.W. The HSV-1 mechanisms of cell-to-cell spread and fusion are critically dependent on host PTP1B. PLoS Pathog. 2018, 14, e1007054. [Google Scholar] [CrossRef] [PubMed]
- Ni, L.; Li, Y.; Wu, K.; Deng, F.; Wang, H.; Ning, Y.J. Antitumor efficacy of CRISPR/Cas9-engineered ICP6 mutant herpes simplex viruses in a mouse xenograft model for lung adenocarcinoma. J. Med. Virol. 2022, 94, 6000–6015. [Google Scholar] [CrossRef] [PubMed]
- Shibata, T.; Uchida, H.; Shiroyama, T.; Okubo, Y.; Suzuki, T.; Ikeda, H.; Yamaguchi, M.; Miyagawa, Y.; Fukuhara, T.; Cohen, J.B.; et al. Development of an oncolytic HSV vector fully retargeted specifically to cellular EpCAM for virus entry and cell-to-cell spread. Gene Ther. 2016, 23, 479–488. [Google Scholar] [CrossRef] [PubMed]
- Mateo, M.; Generous, A.; Sinn, P.L.; Cattaneo, R. Connections matter—How viruses use cell–cell adhesion components. J. Cell Sci. 2015, 128, 431–439. [Google Scholar] [CrossRef] [PubMed]
- Greco, A.; Arata, L.; Soler, E.; Gaume, X.; Couté, Y.; Hacot, S.; Callé, A.; Monier, K.; Epstein, A.L.; Sanchez, J.C.; et al. Nucleolin interacts with US11 protein of herpes simplex virus 1 and is involved in its trafficking. J. Virol. 2012, 86, 1449–1457. [Google Scholar] [CrossRef]
- Xing, J.; Wang, S.; Lin, R.; Mossman, K.L.; Zheng, C. Herpes simplex virus 1 tegument protein US11 downmodulates the RLR signaling pathway via direct interaction with RIG-I and MDA-5. J. Virol. 2012, 86, 3528–3540. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Main, D.; Ma, Y.; He, B. Herpes Simplex Virus 1 Inhibits TANK-Binding Kinase 1 through Formation of the Us11-Hsp90 Complex. J. Virol. 2018, 92, 10–128. [Google Scholar] [CrossRef] [PubMed]
- Connolly, S.A.; Jardetzky, T.S.; Longnecker, R. The structural basis of herpesvirus entry. Nat. Rev. Microbiol. 2021, 19, 110–121. [Google Scholar] [CrossRef] [PubMed]
- Ushijima, Y.; Luo, C.; Kamakura, M.; Goshima, F.; Kimura, H.; Nishiyama, Y. Herpes simplex virus UL56 interacts with and regulates the Nedd4-family ubiquitin ligase Itch. Virol. J. 2010, 7, 179. [Google Scholar] [CrossRef] [PubMed]
- Ushijima, Y.; Goshima, F.; Kimura, H.; Nishiyama, Y. Herpes simplex virus type 2 tegument protein UL56 relocalizes ubiquitin ligase Nedd4 and has a role in transport and/or release of virions. Virol. J. 2009, 6, 168. [Google Scholar] [CrossRef]
- Balan, P.; Davis-Poynter, N.; Bell, S.; Atkinson, H.; Browne, H.; Minson, T. An analysis of the in vitro and in vivo phenotypes of mutants of herpes simplex virus type 1 lacking glycoproteins gG, gE, gI or the putative gJ. J. Gen. Virol. 1994, 75 Pt 6, 1245–1258. [Google Scholar] [CrossRef] [PubMed]
- Koshizuka, T.; Kobayashi, T.; Ishioka, K.; Suzutani, T. Herpesviruses possess conserved proteins for interaction with Nedd4 family ubiquitin E3 ligases. Sci. Rep. 2018, 8, 4447. [Google Scholar] [CrossRef] [PubMed]


| Sample No. | Strain Code | NVRC Accession Number | Subject | Site | Province | Date |
|---|---|---|---|---|---|---|
| 1 | HB94 | CSTR:16533.06.IVCAS 6.0180 | N/A | N/A | Hubei | 1994 |
| 2 | HN19 | CSTR:16533.06.IVCAS 6.7531 | Patient female 40 | Lip | Hainan | 2019 |
| Strain Code | No. of Reads Used for Assembly | Avg coverage | Genome Length | Accession No. | Avg ORF Identity with HB94 (nucl/aa) | Avg ORF Identity with HN19 (nucl/aa) | Avg ORF Identity with ZW6 (nucl/aa) | Avg ORF Identity with CR38 (nucl/aa) |
|---|---|---|---|---|---|---|---|---|
| HB94 a | 46.4 × 106 | 46278 X | 151,002 | NAAADWDO10000000 | 100/100 | 99.1/98.9 | 99.3/98.5 | 99.1/98.3 |
| HN19 a | 7.8 × 106 | 7623 X | 152,073 | NAAADWEO10000000 | 99.1/98.9 | 100/100 | 99.6/99.5 | 99.6/99.3 |
| ZW6 b | N/A | N/A | 152,090 | KX424525 | 99.3/98.5 | 99.6/99.5 | 100/100 | 99.6/99.2 |
| CR38 b | N/A | N/A | 151,427 | HM585508 | 99.1/98.3 | 99.6/99.3 | 99.6/99.2 | 100/100 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Wu, K.; Ni, L.; Li, C.; Peng, R.; Li, Y.; Fan, Z.; Yin, F.; Deng, F.; Shen, S.; et al. Effects of US7 and UL56 on Cell-to-Cell Spread of Human Herpes Simplex Virus 1. Viruses 2023, 15, 2256. https://doi.org/10.3390/v15112256
Wang J, Wu K, Ni L, Li C, Peng R, Li Y, Fan Z, Yin F, Deng F, Shen S, et al. Effects of US7 and UL56 on Cell-to-Cell Spread of Human Herpes Simplex Virus 1. Viruses. 2023; 15(11):2256. https://doi.org/10.3390/v15112256
Chicago/Turabian StyleWang, Jun, Ke Wu, Longquan Ni, Chenxuan Li, Ruoyan Peng, Yi Li, Zhaojun Fan, Feifei Yin, Fei Deng, Shu Shen, and et al. 2023. "Effects of US7 and UL56 on Cell-to-Cell Spread of Human Herpes Simplex Virus 1" Viruses 15, no. 11: 2256. https://doi.org/10.3390/v15112256





