Emergence of Intergenogroup Reassortant G9P[4] Strains Following Rotavirus Vaccine Introduction in Ghana
Abstract
:1. Introduction
2. Materials and Methods
2.1. Specimens and Ethical Approval
2.2. RNA Extraction
2.3. cDNA Library Preparation and Illumina MiSeq Sequencing
2.4. Genetic Analysis Algorithms
2.5. Nucleotide Sequence Accession Numbers
3. Results
3.1. The Collection of Samples, Age Distribution, and Proportion of Rotavirus Detection
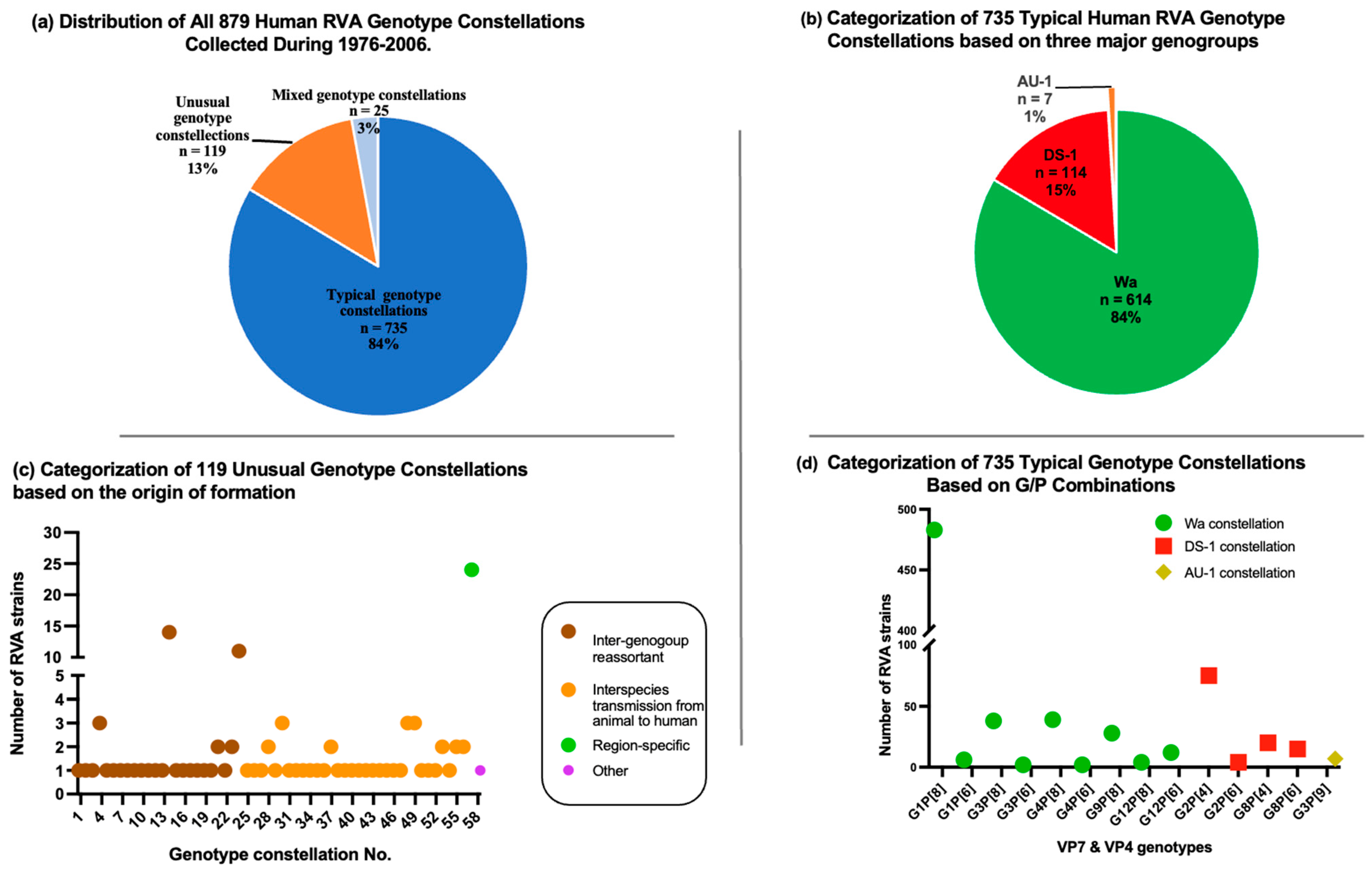
3.2. Genotype Constellation
3.3. Comprehensive Collection of RVA Genotype Constellations in the Pre-Vaccination Era
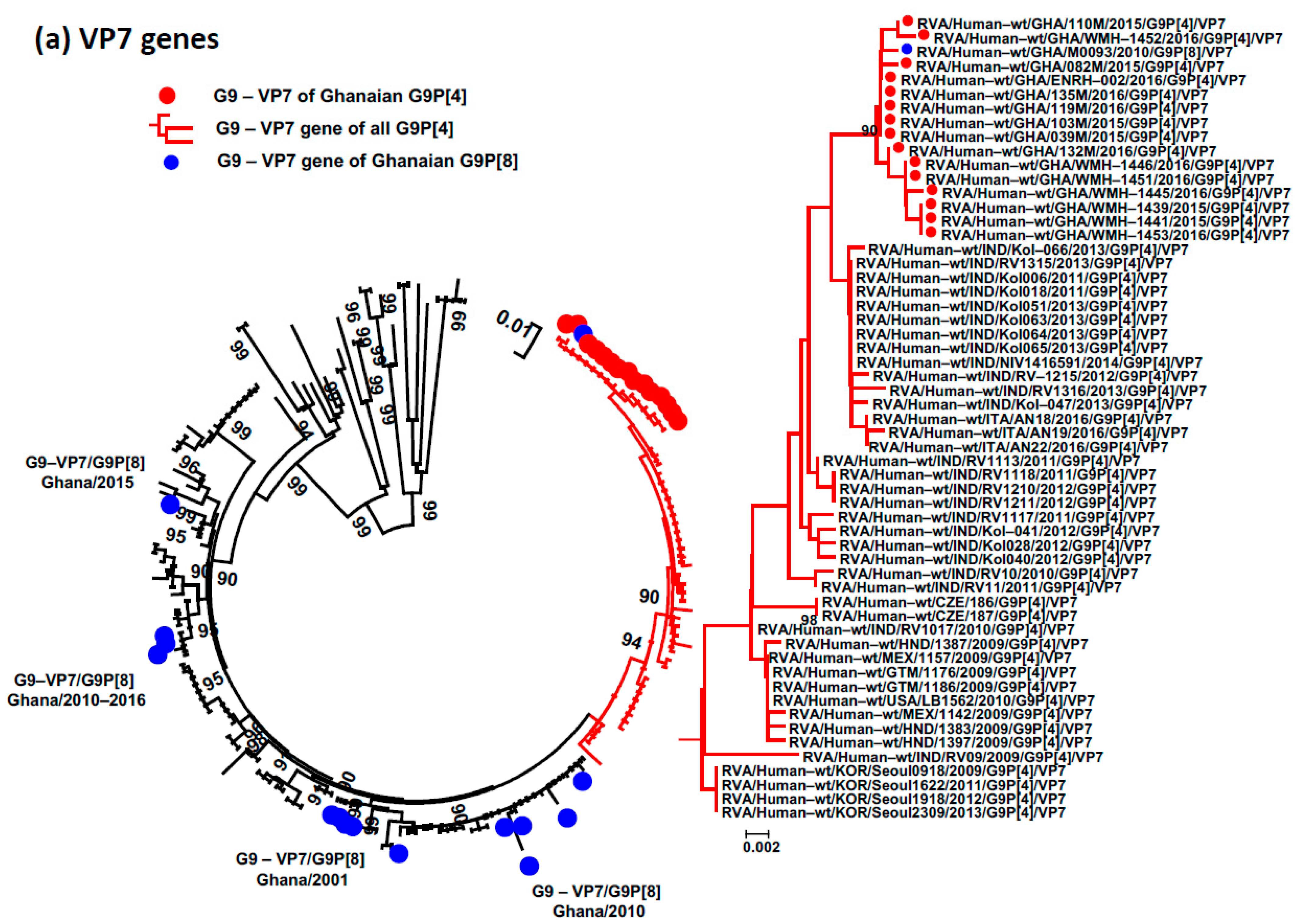
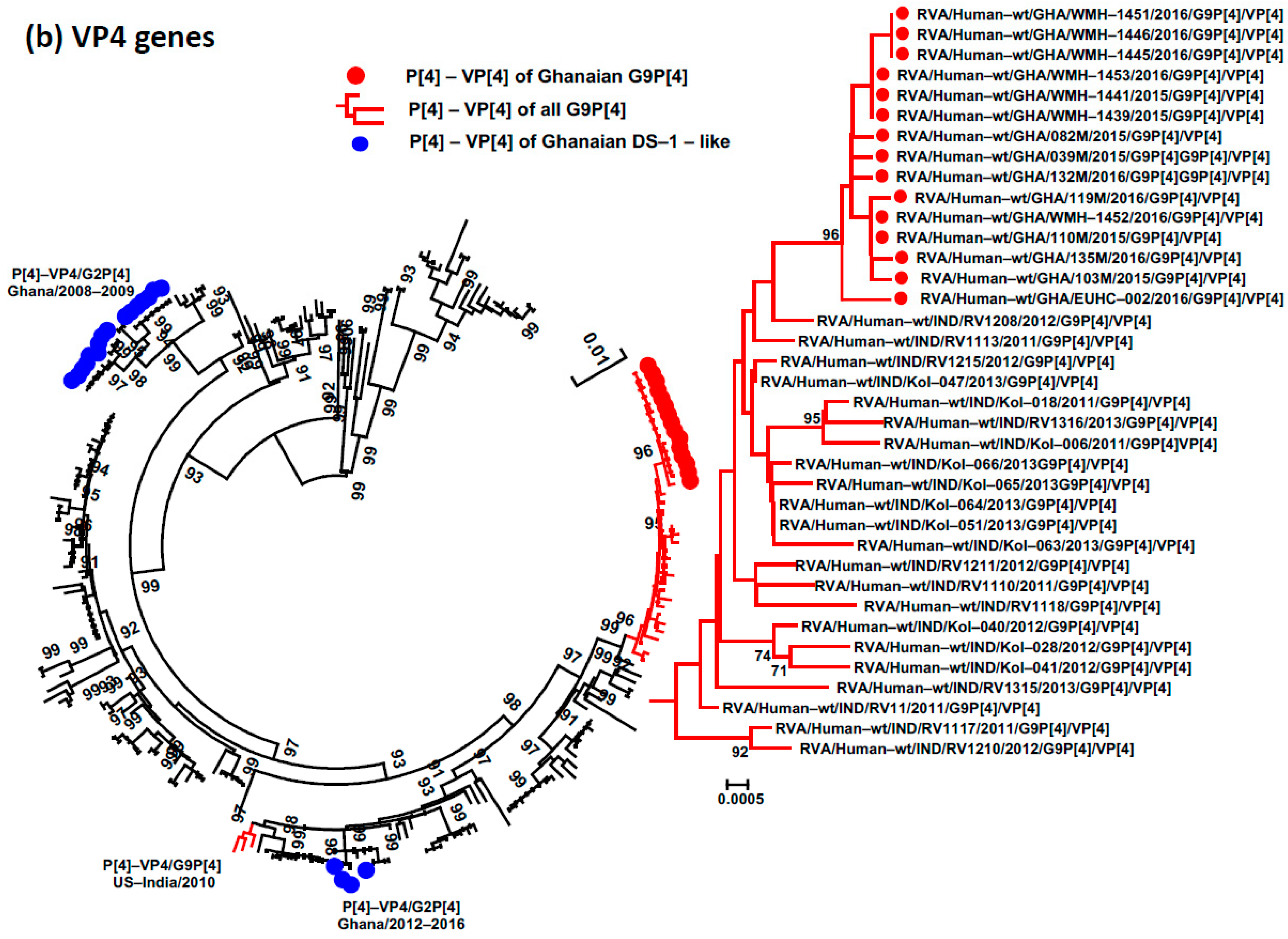
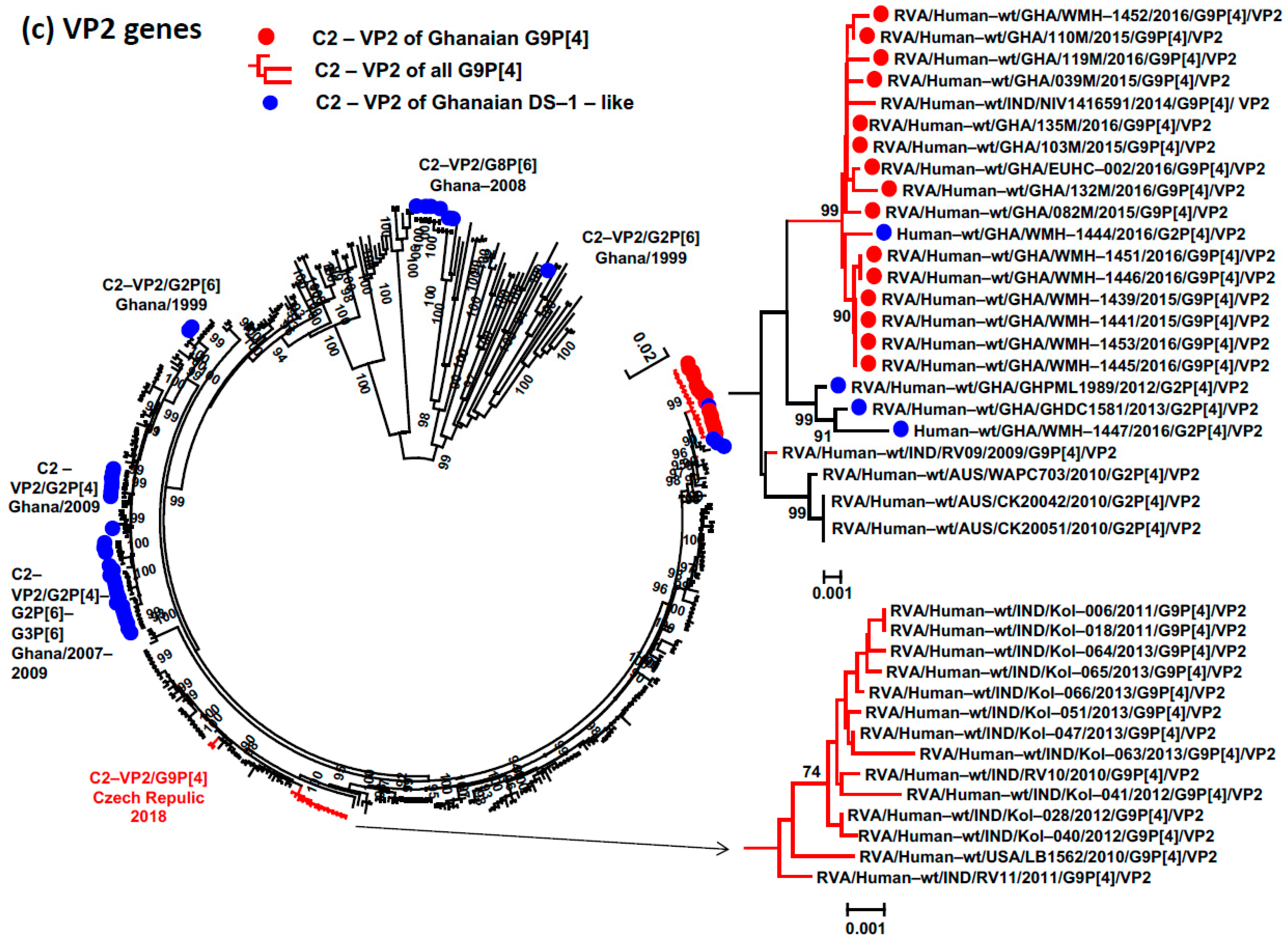
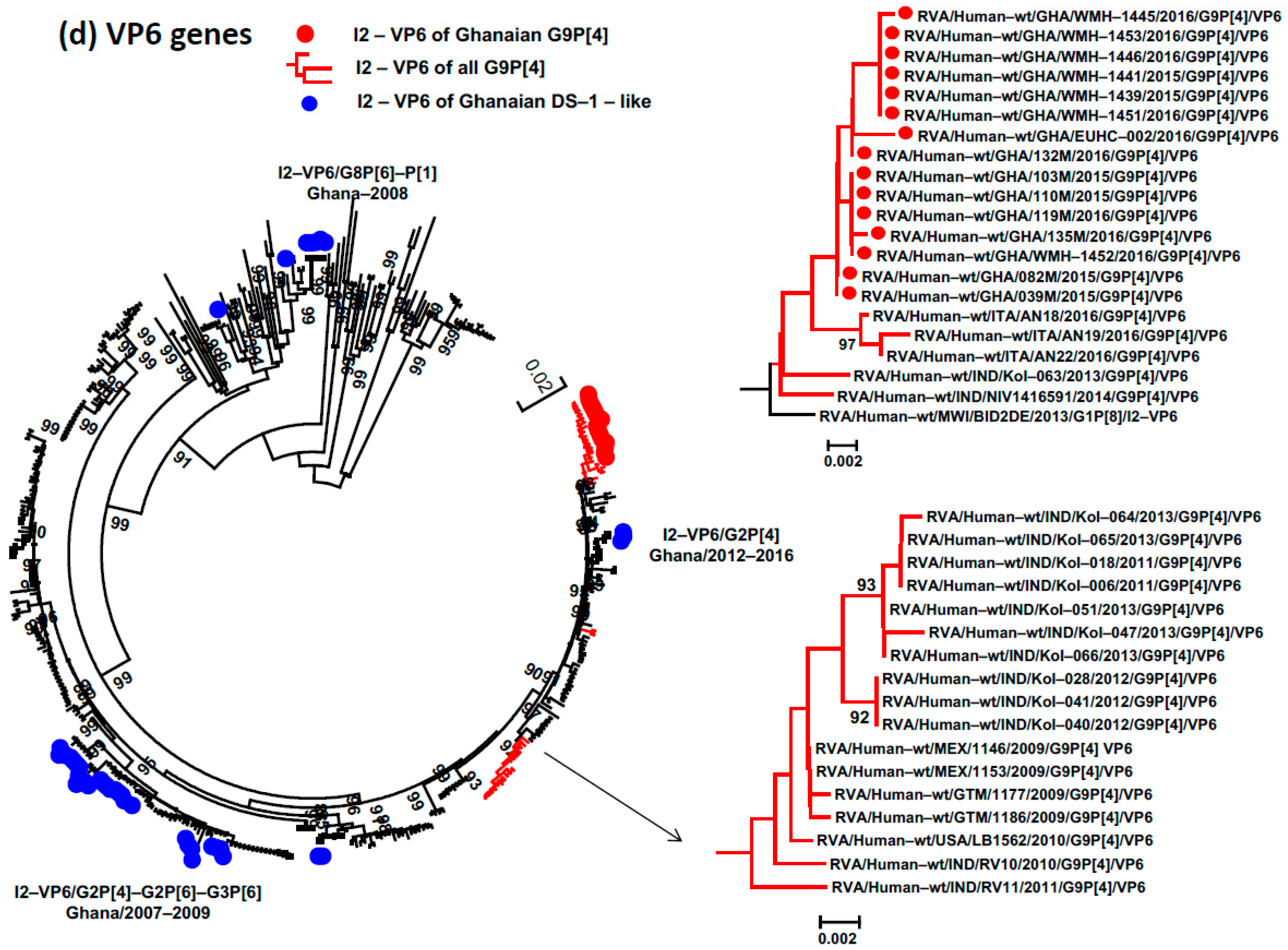
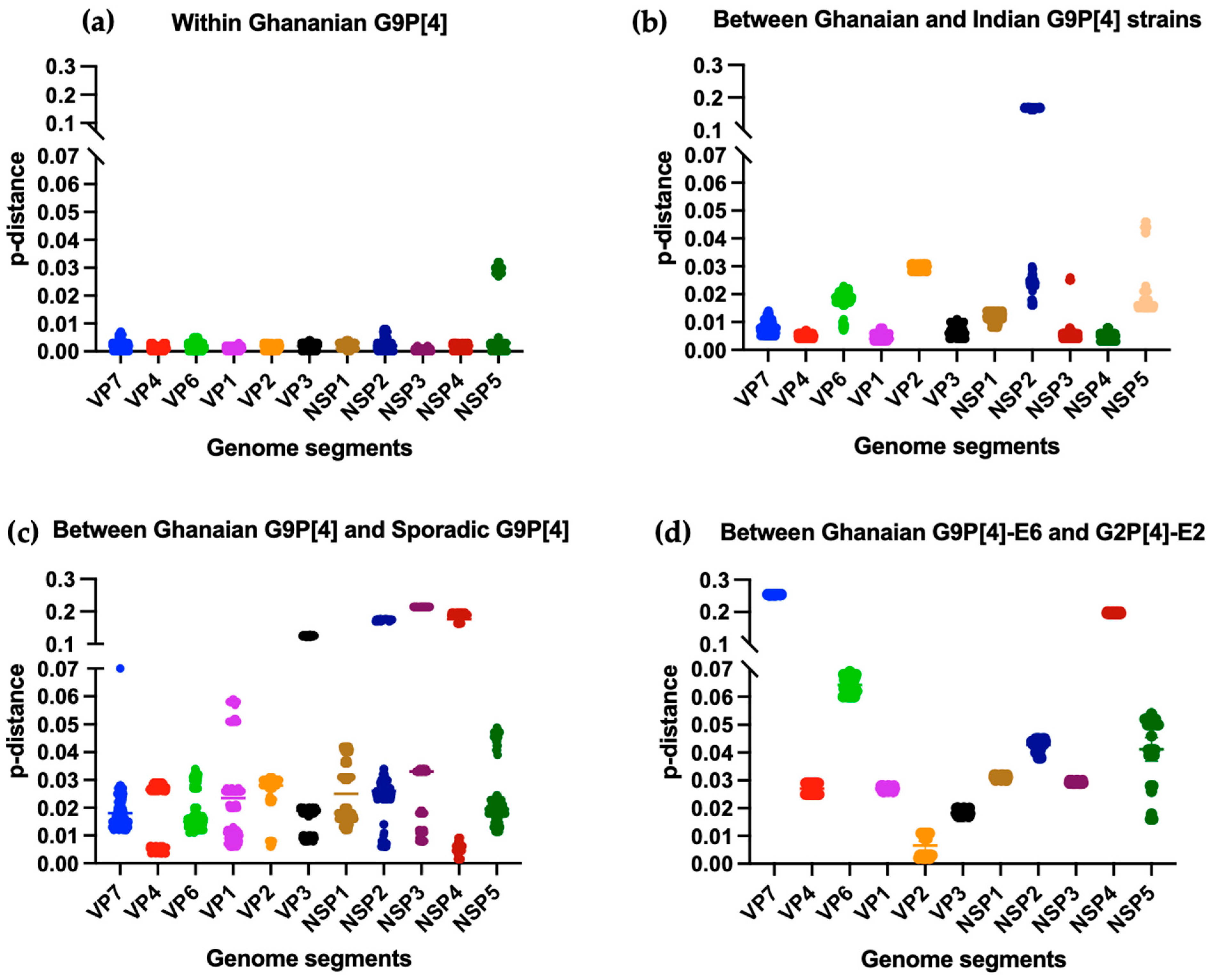
3.4. Phylogenetic Analysis and Proportional p-Distances
3.5. The Circulation of the Uncommon E6 Genotype of the NSP4 Gene
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Parashar, U.D.; Gibson, C.J.; Bresee, J.S.; Glass, R.I. Rotavirus and severe childhood diarrhea. Emerg. Infect. Dis. 2006, 12, 304–306. [Google Scholar] [CrossRef] [PubMed]
- Tate, J.E.; Burton, A.H.; Boschi-Pinto, C.; Parashar, U.D. World Health Organization-Coordinated Global Rotavirus Surveillance, N. Global, Regional, and National Estimates of Rotavirus Mortality in Children < 5 Years of Age, 2000–2013. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2016, 62 (Suppl. S2), S96–S105. [Google Scholar] [CrossRef]
- Tate, J.E.; Burton, A.H.; Boschi-Pinto, C.; Steele, A.D.; Duque, J.; Parashar, U.D.; WHO-coordinated Global Rotavirus Surveillance Network. 2008 estimate of worldwide rotavirus-associated mortality in children younger than 5 years before the introduction of universal rotavirus vaccination programmes: A systematic review and meta-analysis. Lancet. Infect. Dis. 2012, 12, 136–141. [Google Scholar] [CrossRef] [PubMed]
- The Virus Classification and Taxon Nomenclature (ICTV). Available online: https://ictv.global/report/chapter/sedoreoviridae/sedoreoviridae/rotavirus (accessed on 7 December 2023).
- Matthijnssens, J.; Ciarlet, M.; Heiman, E.; Arijs, I.; Delbeke, T.; McDonald, S.M.; Palombo, E.A.; Iturriza-Gomara, M.; Maes, P.; Patton, J.T.; et al. Full genome-based classification of rotaviruses reveals a common origin between human Wa-Like and porcine rotavirus strains and human DS-1-like and bovine rotavirus strains. J. Virol. 2008, 82, 3204–3219. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization Home Page. Available online: https://www.who.int/teams/immunization-vaccines-and-biologicals/diseases/rotavirus (accessed on 7 December 2023).
- Ruiz-Palacios, G.M.; Perez-Schael, I.; Velazquez, F.R.; Abate, H.; Breuer, T.; Clemens, S.C.; Cheuvart, B.; Espinoza, F.; Gillard, P.; Innis, B.L.; et al. Safety and efficacy of an attenuated vaccine against severe rotavirus gastroenteritis. N. Engl. J. Med. 2006, 354, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Vesikari, T.; Matson, D.O.; Dennehy, P.; Van Damme, P.; Santosham, M.; Rodriguez, Z.; Dallas, M.J.; Heyse, J.F.; Goveia, M.G.; Black, S.B.; et al. Safety and efficacy of a pentavalent human-bovine (WC3) reassortant rotavirus vaccine. N. Engl. J. Med. 2006, 354, 23–33. [Google Scholar] [CrossRef] [PubMed]
- Troeger, C.; Khalil, I.A.; Rao, P.C.; Cao, S.; Blacker, B.F.; Ahmed, T.; Armah, G.; Bines, J.E.; Brewer, T.G.; Colombara, D.V.; et al. Rotavirus Vaccination and the Global Burden of Rotavirus Diarrhea Among Children Younger than 5 Years. JAMA Pediatr. 2018, 172, 958–965. [Google Scholar] [CrossRef] [PubMed]
- Fujii, Y.; Nakagomi, T.; Nishimura, N.; Noguchi, A.; Miura, S.; Ito, H.; Doan, Y.H.; Takahashi, T.; Ozaki, T.; Katayama, K.; et al. Spread and predominance in Japan of novel G1P[8] double-reassortant rotavirus strains possessing a DS-1-like genotype constellation typical of G2P[4] strains. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2014, 28, 426–433. [Google Scholar] [CrossRef]
- Hoa-Tran, T.N.; Nakagomi, T.; Vu, H.M.; Do, L.P.; Gauchan, P.; Agbemabiese, C.A.; Nguyen, T.T.; Nakagomi, O.; Thanh, N.T. Abrupt emergence and predominance in Vietnam of rotavirus A strains possessing a bovine-like G8 on a DS-1-like background. Arch. Virol. 2016, 161, 479–482. [Google Scholar] [CrossRef]
- Komoto, S.; Tacharoenmuang, R.; Guntapong, R.; Ide, T.; Haga, K.; Katayama, K.; Kato, T.; Ouchi, Y.; Kurahashi, H.; Tsuji, T.; et al. Emergence and Characterization of Unusual DS-1-Like G1P[8] Rotavirus Strains in Children with Diarrhea in Thailand. PLoS ONE 2015, 10, e0141739. [Google Scholar] [CrossRef]
- Luchs, A.; da Costa, A.C.; Cilli, A.; Komninakis, S.C.V.; Carmona, R.C.C.; Boen, L.; Morillo, S.G.; Sabino, E.C.; Timenetsky, M. Spread of the emerging equine-like G3P[8] DS-1-like genetic backbone rotavirus strain in Brazil and identification of potential genetic variants. J. Gen. Virol. 2019, 100, 7–25. [Google Scholar] [CrossRef] [PubMed]
- Nakagomi, T.; Nguyen, M.Q.; Gauchan, P.; Agbemabiese, C.A.; Kaneko, M.; Do, L.P.; Vu, T.D.; Nakagomi, O. Evolution of DS-1-like G1P[8] double-gene reassortant rotavirus A strains causing gastroenteritis in children in Vietnam in 2012/2013. Arch. Virol. 2017, 162, 739–748. [Google Scholar] [CrossRef] [PubMed]
- Katz, E.M.; Esona, M.D.; Betrapally, N.S.; De La Cruz De Leon, L.A.; Neira, Y.R.; Rey, G.J.; Bowen, M.D. Whole-gene analysis of inter-genogroup reassortant rotaviruses from the Dominican Republic: Emergence of equine-like G3 strains and evidence of their reassortment with locally-circulating strains. Virology 2019, 534, 114–131. [Google Scholar] [CrossRef] [PubMed]
- Wandera, E.A.; Komoto, S.; Mohammad, S.; Ide, T.; Bundi, M.; Nyangao, J.; Kathiiko, C.; Odoyo, E.; Galata, A.; Miring’u, G.; et al. Genomic characterization of uncommon human G3P[6] rotavirus strains that have emerged in Kenya after rotavirus vaccine introduction, and pre-vaccine human G8P[4] rotavirus strains. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2019, 68, 231–248. [Google Scholar] [CrossRef] [PubMed]
- Cowley, D.; Donato, C.M.; Roczo-Farkas, S.; Kirkwood, C.D. Emergence of a novel equine-like G3P[8] inter-genogroup reassortant rotavirus strain associated with gastroenteritis in Australian children. J. Gen. Virol. 2015, 97, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Arana, A.; Montes, M.; Jere, K.C.; Alkorta, M.; Iturriza-Gomara, M.; Cilla, G. Emergence and spread of G3P[8] rotaviruses possessing an equine-like VP7 and a DS-1-like genetic backbone in the Basque Country (North of Spain), 2015. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2016, 44, 137–144. [Google Scholar] [CrossRef] [PubMed]
- Giri, S.; Kumar, C.P.G.; Khakha, S.A.; Chawla-Sarkar, M.; Gopalkrishna, V.; Chitambar, S.D.; Ray, P.; Venkatasubramanian, S.; Borkakoty, B.J.; Roy, S.; et al. Diversity of rotavirus genotypes circulating in children < 5 years of age hospitalized for acute gastroenteritis in India from 2005 to 2016: Analysis of temporal and regional genotype variation. BMC Infect. Dis. 2020, 20, 740. [Google Scholar] [CrossRef]
- Doan, Y.H.; Suzuki, Y.; Fujii, Y.; Haga, K.; Fujimoto, A.; Takai-Todaka, R.; Someya, Y.; Nayak, M.K.; Mukherjee, A.; Imamura, D.; et al. Complex reassortment events of unusual G9P[4] rotavirus strains in India between 2011 and 2013. Infect. Genet. Evol. 2017, 54, 417–428. [Google Scholar] [CrossRef]
- Quaye, O.; McDonald, S.; Esona, M.D.; Lyde, F.C.; Mijatovic-Rustempasic, S.; Roy, S.; Banegas, D.J.; Quinonez, Y.M.; Chinchilla, B.L.; Santiago, F.G.; et al. Rotavirus G9P[4] in 3 countries in Latin America, 2009–2010. Emerg. Infect. Dis. 2013, 19, 1332–1333. [Google Scholar] [CrossRef]
- Afrad, M.H.; Rahman, M.Z.; Matthijnssens, J.; Das, S.K.; Faruque, A.S.; Azim, T.; Rahman, M. High incidence of reassortant G9P[4] rotavirus strain in Bangladesh: Fully heterotypic from vaccine strains. J. Clin. Virol. 2013, 58, 755–756. [Google Scholar] [CrossRef]
- Lewis, J.; Roy, S.; Esona, M.D.; Mijatovic-Rustempasic, S.; Hardy, C.; Wang, Y.; Cortese, M.; Bowen, M.D. Full Genome Sequence of a Reassortant Human G9P[4] Rotavirus Strain. Genome Announc. 2014, 2, 10–128. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, S.P.; Kaida, A.; Ono, A.; Kubo, H.; Iritani, N. Detection and characterization of a human G9P[4] rotavirus strain in Japan. J. Med. Virol. 2015, 87, 1311–1318. [Google Scholar] [CrossRef] [PubMed]
- Ianiro, G.; Recanatini, C.; D’Errico, M.M.; Monini, M.; RotaNet-Italy Study, G. Uncommon G9P[4] group A rotavirus strains causing dehydrating diarrhea in young children in Italy. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2018, 64, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Moutelikova, R.; Sauer, P.; Prodelalova, J. Whole-genome sequence of a reassortant G9P[4] rotavirus A strain from two children in the Czech Republic. Arch. Virol. 2020, 165, 1703–1706. [Google Scholar] [CrossRef] [PubMed]
- Lartey, B.L.; Damanka, S.; Dennis, F.E.; Enweronu-Laryea, C.C.; Addo-Yobo, E.; Ansong, D.; Kwarteng-Owusu, S.; Sagoe, K.W.; Mwenda, J.M.; Diamenu, S.K.; et al. Rotavirus strain distribution in Ghana pre- and post- rotavirus vaccine introduction. Vaccine 2018, 36, 7238–7242. [Google Scholar] [CrossRef] [PubMed]
- Gentsch, J.R.; Glass, R.I.; Woods, P.; Gouvea, V.; Gorziglia, M.; Flores, J.; Das, B.K.; Bhan, M.K. Identification of group A rotavirus gene 4 types by polymerase chain reaction. J. Clin. Microbiol. 1992, 30, 1365–1373. [Google Scholar] [CrossRef]
- Pickett, B.E.; Greer, D.S.; Zhang, Y.; Stewart, L.; Zhou, L.; Sun, G.; Gu, Z.; Kumar, S.; Zaremba, S.; Larsen, C.N.; et al. Virus pathogen database and analysis resource (ViPR): A comprehensive bioinformatics database and analysis resource for the coronavirus research community. Viruses 2012, 4, 3209–3226. [Google Scholar] [CrossRef] [PubMed]
- Agbemabiese, C.A.; Nakagomi, T.; Damanka, S.A.; Dennis, F.E.; Lartey, B.L.; Armah, G.E.; Nakagomi, O. Sub-genotype phylogeny of the non-G, non-P genes of genotype 2 Rotavirus A strains. PLoS ONE 2019, 14, e0217422. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Doan, Y.H.; Nakagomi, T.; Agbemabiese, C.A.; Nakagomi, O. Changes in the distribution of lineage constellations of G2P[4] Rotavirus A strains detected in Japan over 32 years (1980–2011). Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2015, 34, 423–433. [Google Scholar] [CrossRef]
- Doan, Y.H.; Nakagomi, T.; Nakagomi, O. Repeated circulation over 6 years of intergenogroup mono-reassortant G2P[4] rotavirus strains with genotype N1 of the NSP2 gene. Infect. Genet. Evol. 2012, 12, 1202–1212. [Google Scholar] [CrossRef] [PubMed]
- Armah, G.; Pringle, K.; Enweronu-Laryea, C.C.; Ansong, D.; Mwenda, J.M.; Diamenu, S.K.; Narh, C.; Lartey, B.; Binka, F.; Grytdal, S.; et al. Impact and Effectiveness of Monovalent Rotavirus Vaccine Against Severe Rotavirus Diarrhea in Ghana. Clin. Infect. Dis. 2016, 62 (Suppl. S2), S200–S207. [Google Scholar] [CrossRef] [PubMed]
- Araujo, I.T.; Ferreira, M.S.; Fialho, A.M.; Assis, R.M.; Cruz, C.M.; Rocha, M.; Leite, J.P. Rotavirus genotypes P[4]G9, P[6]G9, and P[8]G9 in hospitalized children with acute gastroenteritis in Rio de Janeiro, Brazil. J. Clin. Microbiol. 2001, 39, 1999–2001. [Google Scholar] [CrossRef] [PubMed]
- Santos, N.; Volotao, E.M.; Soares, C.C.; Albuquerque, M.C.; da Silva, F.M.; de Carvalho, T.R.; Pereira, C.F.; Chizhikov, V.; Hoshino, Y. Rotavirus strains bearing genotype G9 or P[9] recovered from Brazilian children with diarrhea from 1997 to 1999. J. Clin. Microbiol. 2001, 39, 1157–1160. [Google Scholar] [CrossRef]
- Esona, M.D.; Armah, G.E.; Steele, A.D. Rotavirus VP4 and VP7 genotypes circulating in Cameroon: Identification of unusual types. J. Infect. Dis. 2010, 202 (Suppl. S1), S205–S211. [Google Scholar] [CrossRef]
- Zeller, M.; Donato, C.; Trovao, N.S.; Cowley, D.; Heylen, E.; Donker, N.C.; McAllen, J.K.; Akopov, A.; Kirkness, E.F.; Lemey, P.; et al. Genome-Wide Evolutionary Analyses of G1P[8] Strains Isolated Before and After Rotavirus Vaccine Introduction. Genome Biol. Evol. 2015, 7, 2473–2483. [Google Scholar] [CrossRef]
- da Silva, M.F.M.; Rose, T.L.; Gomez, M.M.; Carvalho-Costa, F.A.; Fialho, A.M.; de Assis, R.M.S.; de Andrade, J.; Volotao, E.M.; Leite, J.P.G. G1P[8] species A rotavirus over 27 years--pre- and post-vaccination eras--in Brazil: Full genomic constellation analysis and no evidence for selection pressure by Rotarix(R) vaccine. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2015, 30, 206–218. [Google Scholar] [CrossRef]
- Munlela, B.; Joao, E.D.; Donato, C.M.; Strydom, A.; Boene, S.S.; Chissaque, A.; Bauhofer, A.F.L.; Langa, J.; Cassocera, M.; Cossa-Moiane, I.; et al. Whole Genome Characterization and Evolutionary Analysis of G1P[8] Rotavirus A Strains during the Pre- and Post-Vaccine Periods in Mozambique (2012–2017). Pathogens 2020, 9, 26. [Google Scholar] [CrossRef]
- Mwangi, P.N.; Mogotsi, M.T.; Seheri, M.L.; Mphahlele, M.J.; Peenze, I.; Esona, M.D.; Kumwenda, B.; Steele, A.D.; Kirkwood, C.D.; Ndze, V.N.; et al. Whole Genome In-Silico Analysis of South African G1P[8] Rotavirus Strains Before and After Vaccine Introduction Over A Period of 14 Years. Vaccines 2020, 8, 609. [Google Scholar] [CrossRef]
- Rasebotsa, S.; Mwangi, P.N.; Mogotsi, M.T.; Sabiu, S.; Magagula, N.B.; Rakau, K.; Uwimana, J.; Mutesa, L.; Muganga, N.; Murenzi, D.; et al. Whole genome and in-silico analyses of G1P[8] rotavirus strains from pre- and post-vaccination periods in Rwanda. Sci. Rep. 2020, 10, 13460. [Google Scholar] [CrossRef]
- Zhang, S.; McDonald, P.W.; Thompson, T.A.; Dennis, A.F.; Akopov, A.; Kirkness, E.F.; Patton, J.T.; McDonald, S.M. Analysis of human rotaviruses from a single location over an 18-year time span suggests that protein coadaption influences gene constellations. J. Virol. 2014, 88, 9842–9863. [Google Scholar] [CrossRef]
- Nakagomi, T.; Cuevas, L.E.; Gurgel, R.G.; Elrokhsi, S.H.; Belkhir, Y.A.; Abugalia, M.; Dove, W.; Montenegro, F.M.; Correia, J.B.; Nakagomi, O.; et al. Apparent extinction of non-G2 rotavirus strains from circulation in Recife, Brazil, after the introduction of rotavirus vaccine. Arch. Virol. 2008, 153, 591–593. [Google Scholar] [CrossRef]
- Kirkwood, C.D.; Boniface, K.; Barnes, G.L.; Bishop, R.F. Distribution of rotavirus genotypes after introduction of rotavirus vaccines, Rotarix(R) and RotaTeq(R), into the National Immunization Program of Australia. Pediatr. Infect. Dis. J. 2011, 30, S48–S53. [Google Scholar] [CrossRef]
- Matthijnssens, J.; Zeller, M.; Heylen, E.; De Coster, S.; Vercauteren, J.; Braeckman, T.; Van Herck, K.; Meyer, N.; Pircon, J.Y.; Soriano-Gabarro, M.; et al. Higher proportion of G2P[4] rotaviruses in vaccinated hospitalized cases compared with unvaccinated hospitalized cases, despite high vaccine effectiveness against heterotypic G2P[4] rotaviruses. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2014, 20, O702–O710. [Google Scholar] [CrossRef]
- Jere, K.C.; Bar-Zeev, N.; Chande, A.; Bennett, A.; Pollock, L.; Sanchez-Lopez, P.F.; Nakagomi, O.; Tate, J.E.; Parashar, U.D.; Heyderman, R.S.; et al. Vaccine Effectiveness against DS-1-Like Rotavirus Strains in Infants with Acute Gastroenteritis, Malawi, 2013-2015. Emerg. Infect. Dis. 2019, 25, 1734–1737. [Google Scholar] [CrossRef]
- Yen, C.; Figueroa, J.R.; Uribe, E.S.; Carmen-Hernandez, L.D.; Tate, J.E.; Parashar, U.D.; Patel, M.M.; Richardson Lopez-Collado, V. Monovalent rotavirus vaccine provides protection against an emerging fully heterotypic G9P[4] rotavirus strain in Mexico. J. Infect. Dis. 2011, 204, 783–786. [Google Scholar] [CrossRef]
- Sharma, S.; Paul, V.K.; Bhan, M.K.; Ray, P. Genomic characterization of nontypeable rotaviruses and detection of a rare G8 strain in Delhi, India. J. Clin. Microbiol. 2009, 47, 3998–4005. [Google Scholar] [CrossRef]
- Rahman, M.; Matthijnssens, J.; Yang, X.; Delbeke, T.; Arijs, I.; Taniguchi, K.; Iturriza-Gomara, M.; Iftekharuddin, N.; Azim, T.; Van Ranst, M. Evolutionary history and global spread of the emerging g12 human rotaviruses. J. Virol. 2007, 81, 2382–2390. [Google Scholar] [CrossRef]
- De Grazia, S.; Giammanco, G.M.; Doro, R.; Bonura, F.; Marton, S.; Cascio, A.; Martella, V.; Banyai, K. Identification of a multi-reassortant G12P[9] rotavirus with novel VP1, VP2, VP3 and NSP2 genotypes in a child with acute gastroenteritis. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2015, 35, 34–37. [Google Scholar] [CrossRef]
- Kachooei, A.; Tava Koli, A.; Minaeian, S.; Hosseini, M.; Jalilvand, S.; Latifi, T.; Arashkia, A.; Ataei-Pirkooh, A.; Shoja, Z. Molecular characterization of rotavirus infections in children less than 5 years of age with acute gastroenteritis in Tehran, Iran, 2021-2022: Emergence of uncommon G9P[4] and G9P[8] rotavirus strains. J. Med. Virol. 2023, 95, e28529. [Google Scholar] [CrossRef]
- Athiyyah, A.F.; Utsumi, T.; Wahyuni, R.M.; Dinana, Z.; Yamani, L.N.; Soetjipto; Sudarmo, S.M.; Ranuh, R.G.; Darma, A.; Juniastuti; et al. Molecular Epidemiology and Clinical Features of Rotavirus Infection Among Pediatric Patients in East Java, Indonesia During 2015–2018: Dynamic Changes in Rotavirus Genotypes from Equine-Like G3 to Typical Human G1/G3. Front. Microbiol. 2019, 10, 940. [Google Scholar] [CrossRef]

| Age Distribution (Months) | Navrongo | Greater Accra | Western Region | Total | ||||
|---|---|---|---|---|---|---|---|---|
| Number of AGE Samples | Number of RVA Samples (%) | Number of AGE Samples | Number of RVA Samples (%) | Number of AGE Samples | Number of RVA Samples (%) | Number of AGE Samples | Number of RVA Samples (%) | |
| <6 | 3 | 0 (0) | 129 | 26 (20.2) | 10 | 3 (30.0) | 142 | 29 (17.1) |
| 6–12 | 30 | 10 (33.3) | 162 | 36 (22.2) | 48 | 22 (45.8) | 240 | 68 (23.1) |
| 13–24 | 25 | 14 (56.0) | 199 | 43 (21.6) | 60 | 37 (61.7) | 284 | 94 (25.2) |
| >24 | 17 | 3 (17.6) | 93 | 21 (22.6) | 8 | 2 (25.0) | 118 | 26 (21.2) |
| Total | 75 | 27 (36.0) | 583 | 126 (21.6) | 126 | 64 (50.8) | 784 | 217 (27.7) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Doan, Y.H.; Dennis, F.E.; Takemae, N.; Haga, K.; Shimizu, H.; Appiah, M.G.; Lartey, B.L.; Damanka, S.A.; Hayashi, T.; Suzuki, T.; et al. Emergence of Intergenogroup Reassortant G9P[4] Strains Following Rotavirus Vaccine Introduction in Ghana. Viruses 2023, 15, 2453. https://doi.org/10.3390/v15122453
Doan YH, Dennis FE, Takemae N, Haga K, Shimizu H, Appiah MG, Lartey BL, Damanka SA, Hayashi T, Suzuki T, et al. Emergence of Intergenogroup Reassortant G9P[4] Strains Following Rotavirus Vaccine Introduction in Ghana. Viruses. 2023; 15(12):2453. https://doi.org/10.3390/v15122453
Chicago/Turabian StyleDoan, Yen Hai, Francis Ekow Dennis, Nobuhiro Takemae, Kei Haga, Hiroyuki Shimizu, Michael Gyasi Appiah, Belinda Larteley Lartey, Susan Afua Damanka, Takaya Hayashi, Toshihiko Suzuki, and et al. 2023. "Emergence of Intergenogroup Reassortant G9P[4] Strains Following Rotavirus Vaccine Introduction in Ghana" Viruses 15, no. 12: 2453. https://doi.org/10.3390/v15122453







