SARS-CoV-2 Omicron Specific Mutations Affecting Infectivity, Fusogenicity, and Partial TMPRSS2-Independency
Abstract
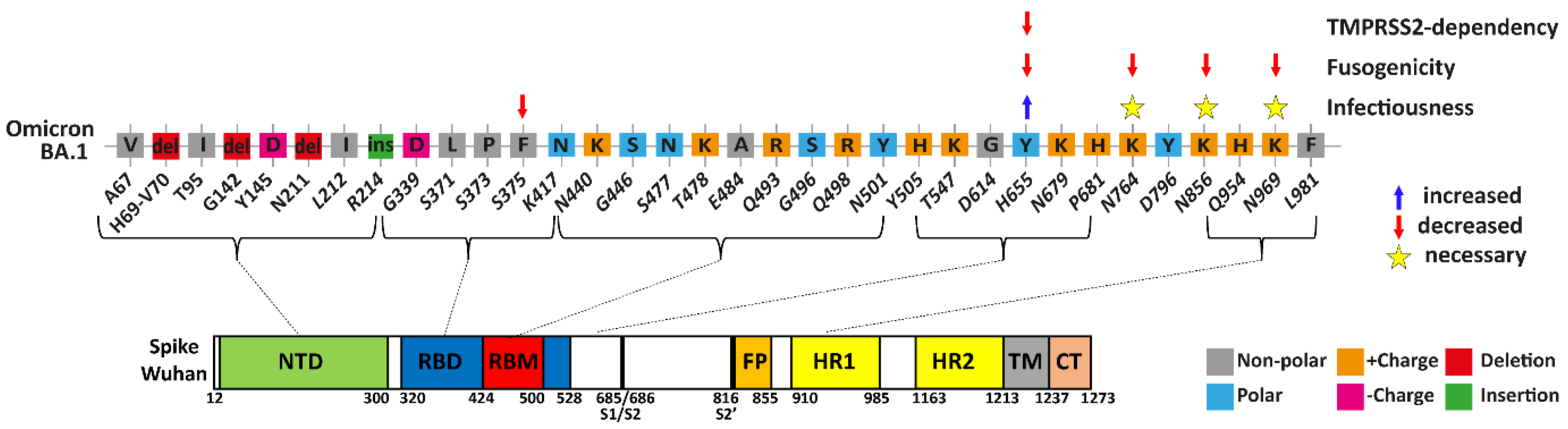
:1. Introduction
2. Methods
2.1. Cell Culture
2.2. Plasmids
2.3. Transfection, Generating of Pseudovirus, and Transduction
2.4. Cell-Fusion Assay
2.5. Graphs and Statistics
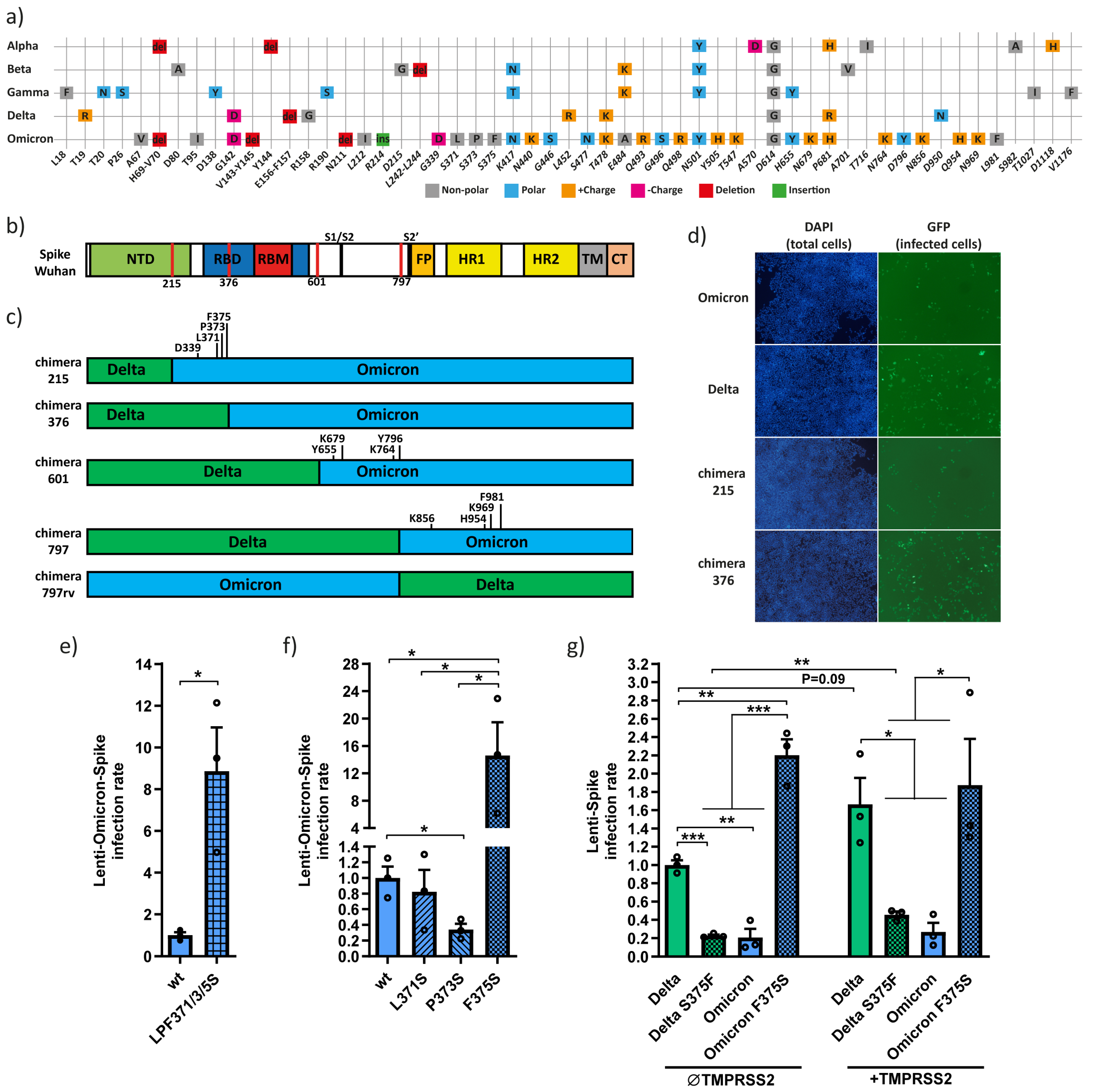
3. Results
3.1. The Omicron F375 Residue Decreases Infection
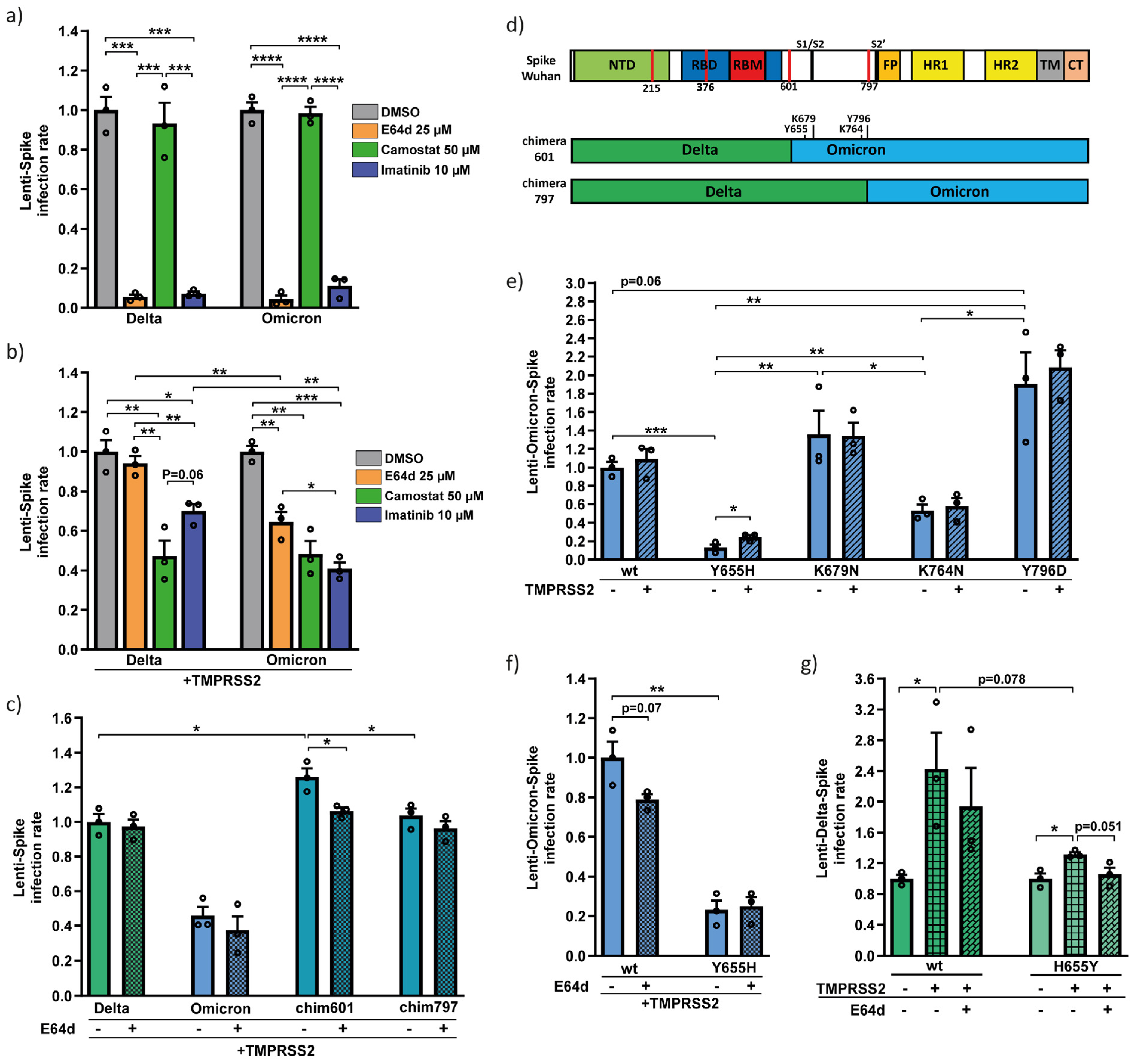
3.2. Omicron Y655 Residue Supports Endosomal Route of Infection
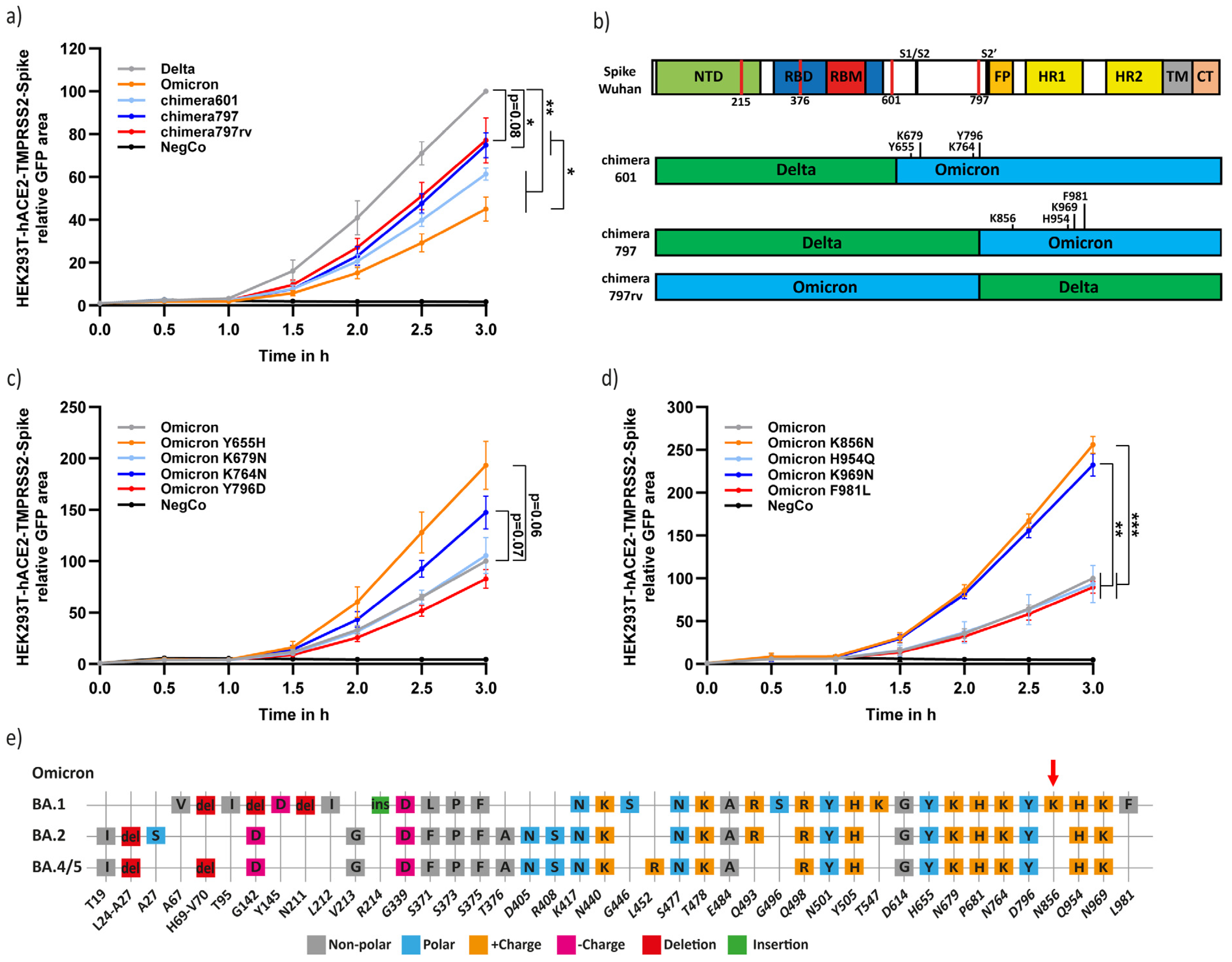
3.3. Residues Y655, K764, K856, and K969 Attenuate Omicron-Spike Membrane Fusion
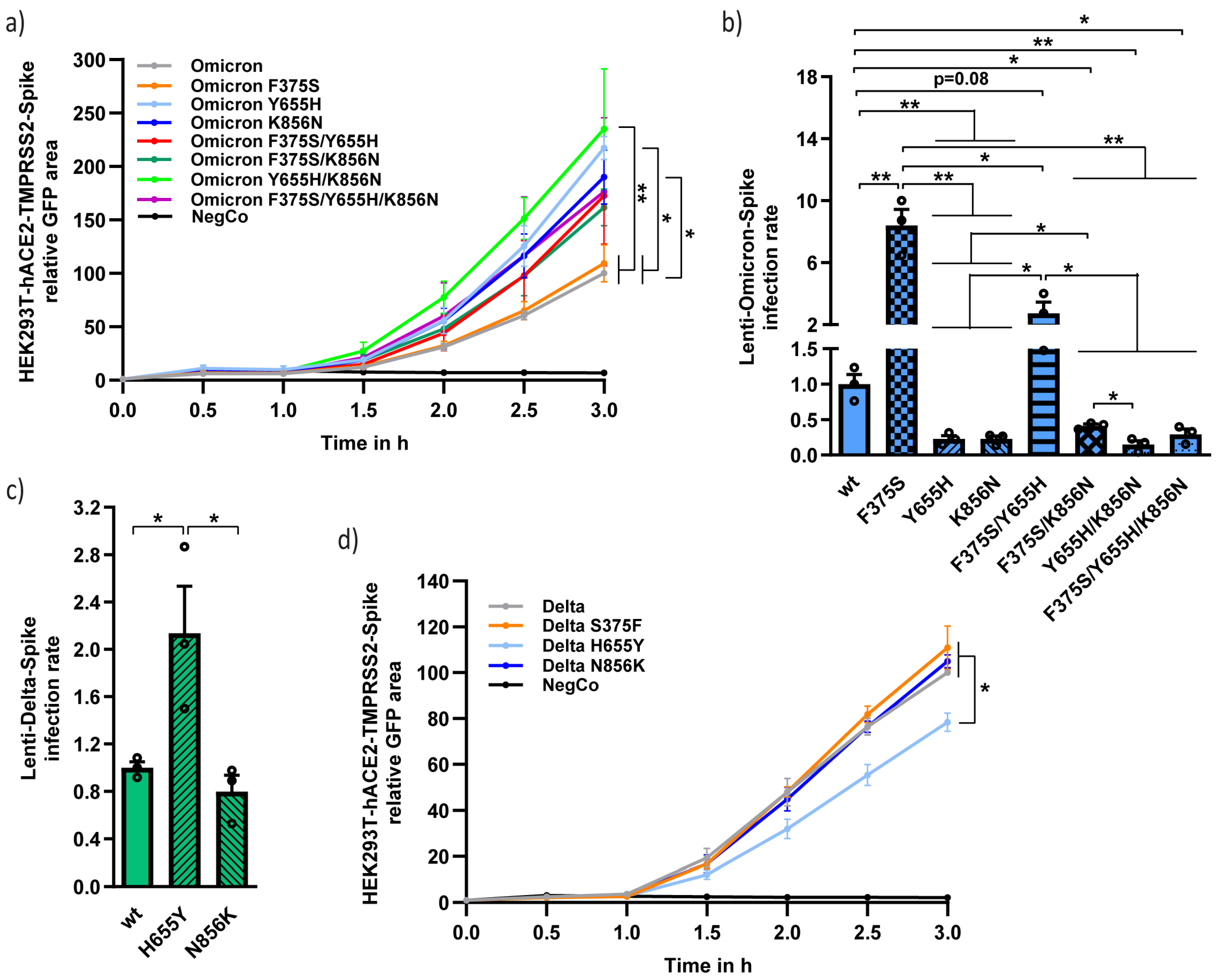
3.4. Functional Analysis of the Different Unique Omicron Spike Residues in Combination
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Horby, P.W.; Hayden, F.G.; Gao, G.F. A novel coronavirus outbreak of global health concern. Lancet 2020, 395, 470–473. [Google Scholar] [CrossRef] [PubMed]
- Gorbalenya, A.E.; Baker, S.C.; Baric, R.S.; de Groot, R.J.; Drosten, C.; Gulyaeva, A.A.; Haagmans, B.L.; Lauber, C.; Leontovich, A.M.; Neuman, B.W.; et al. The Species Severe Acute Respiratory Syndrome-Related Coronavirus: Classifying 2019-NCoV and Naming It SARS-CoV-2. Nat. Microbiol. 2020, 5, 536–544. [Google Scholar]
- World Health Organization (WHO). WHO Tracking SARS-CoV-2 Variants. Available online: https://www.who.int/activities/tracking-SARS-CoV-2-variants (accessed on 28 February 2023).
- Tao, K.; Tzou, P.L.; Nouhin, J.; Gupta, R.K.; de Oliveira, T.; Kosakovsky Pond, S.L.; Fera, D.; Shafer, R.W. The biological and clinical significance of emerging SARS-CoV-2 variants. Nat. Rev. Genet. 2021, 22, 757–773. [Google Scholar] [CrossRef] [PubMed]
- Saito, A.; Irie, T.; Suzuki, R.; Maemura, T.; Nasser, H.; Uriu, K.; Kosugi, Y.; Shirakawa, K.; Sadamasu, K.; Kimura, I.; et al. Enhanced fusogenicity and pathogenicity of SARS-CoV-2 Delta P681R mutation. Nature 2021, 602, 300–306. [Google Scholar] [CrossRef] [PubMed]
- Mlcochova, P.; Kemp, S.A.; Dhar, M.S.; Papa, G.; Meng, B.; Ferreira, I.A.T.M.; Datir, R.; Collier, D.A.; Albecka, A.; Singh, S.; et al. SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion. Nature 2021, 599, 114–119. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization (WHO). Classification of Omicron (B.1.1.529): SARS-CoV-2 Variant of Concern. Available online: https://www.who.int/news/item/26-11-2021-classification-of-omicron-(b.1.1.529)-sars-cov-2-variant-of-concern (accessed on 2 January 2022).
- Network for Genomic Surveillance in South Africa (NGS-SA): SARS-CoV-2 Sequencing Update 26 November 2021; Prepared by the National Institute for Communicable Diseases (NICD) of the National Health Laboratory (NHLS) on Behalf of the Network for Genomics Surveillance in South Africa (NGS-SA). Available online: https://www.nicd.ac.za/wp-content/uploads/2021/11/Update-of-SA-sequencing-data-from-GISAID-26-Nov_Final.pdf (accessed on 3 January 2022).
- Viana, R.; Moyo, S.; Amoako, D.G.; Tegally, H.; Scheepers, C.; Althaus, C.L.; Anyaneji, U.J.; Bester, P.A.; Boni, M.F.; Chand, M.; et al. Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa. Nature 2022, 603, 679–686. [Google Scholar] [CrossRef] [PubMed]
- WHO Weekly Epidemiological Update on COVID-19. 28 December 2021. Available online: https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19---28-december-2021 (accessed on 6 January 2022).
- Lyngse, F.P.; Mortensen, L.H.; Denwood, M.J.; Christiansen, L.E.; Møller, C.H.; Skov, R.L.; Spiess, K.; Fomsgaard, A.; Lassaunière, M.M.; Rasmussen, M.; et al. SARS-CoV-2 Omicron VOC Transmission in Danish Households. medRxiv 2021. [Google Scholar] [CrossRef]
- WHO Weekly Epidemiological Update on COVID-19. 22 February 2023. Available online: https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19---22-february-2023 (accessed on 12 March 2023).
- ECDC-European Center for Disease Control. SARS-CoV-2 Variants of Concern as of 27 October 2021. Available online: https://www.ecdc.europa.eu/en/covid-19/variants-concern (accessed on 10 November 2022).
- Dejnirattisai, W.; Huo, J.; Zhou, D.; Zahradník, J.; Supasa, P.; Liu, C.; Duyvesteyn, H.M.E.; Ginn, H.M.; Mentzer, A.J.; Tuekprakhon, A.; et al. SARS-CoV-2 Omicron-B.1.1.529 Leads to Widespread Escape from Neutralizing Antibody Responses. Cell 2022, 185, 467–484.e15. [Google Scholar] [CrossRef]
- Cele, S.; Jackson, L.; Khoury, D.S.; Khan, K.; Moyo-Gwete, T.; Tegally, H.; San, J.E.; Cromer, D.; Scheepers, C.; Amoako, D.; et al. Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature 2021, 602, 654–656. [Google Scholar] [CrossRef]
- Lupala, C.S.; Ye, Y.; Chen, H.; Su, X.-D.; Liu, H. Mutations on RBD of SARS-CoV-2 Omicron variant result in stronger binding to human ACE2 receptor. Biochem. Biophys. Res. Commun. 2022, 590, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Golcuk, M.; Yildiz, A.; Gur, M. Omicron BA.1 and BA.2 variants increase the interactions of SARS-CoV-2 spike glycoprotein with ACE2. J. Mol. Graph. Model. 2022, 117, 108286. [Google Scholar] [CrossRef] [PubMed]
- Hartenian, E.; Nandakumar, D.; Lari, A.; Ly, M.; Tucker, J.M.; Glaunsinger, B.A. The molecular virology of coronaviruses. J. Biol. Chem. 2020, 295, 12910–12934. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e278. [Google Scholar] [CrossRef] [PubMed]
- Koch, J.; Uckeley, Z.M.; Doldan, P.; Stanifer, M.; Boulant, S.; Lozach, P. TMPRSS2 expression dictates the entry route used by SARS-CoV-2 to infect host cells. EMBO J. 2021, 40, e107821. [Google Scholar] [CrossRef] [PubMed]
- Matsuyama, S.; Nao, N.; Shirato, K.; Kawase, M.; Saito, S.; Takayama, I.; Nagata, N.; Sekizuka, T.; Katoh, H.; Kato, F.; et al. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc. Natl. Acad. Sci. USA 2020, 117, 7001–7003. [Google Scholar] [CrossRef]
- Meng, B.; Abdullahi, A.; Ferreira, I.A.T.M.; Goonawardane, N.; Saito, A.; Kimura, I.; Yamasoba, D.; Gerber, P.P.; Fatihi, S.; Rathore, S.; et al. Altered TMPRSS2 usage by SARS-CoV-2 Omicron impacts infectivity and fusogenicity. Nature 2022, 603, 706–714. [Google Scholar] [CrossRef]
- Willett, B.J.; Grove, J.; MacLean, O.A.; Wilkie, C.; De Lorenzo, G.; Furnon, W.; Cantoni, D.; Scott, S.; Logan, N.; Ashraf, S.; et al. SARS-CoV-2 Omicron is an immune escape variant with an altered cell entry pathway. Nat. Microbiol. 2022, 7, 1161–1179. [Google Scholar] [CrossRef]
- Suzuki, R.; Yamasoba, D.; Kimura, I.; Wang, L.; Kishimoto, M.; Ito, J.; Morioka, Y.; Nao, N.; Nasser, H.; Uriu, K.; et al. Attenuated fusogenicity and pathogenicity of SARS-CoV-2 Omicron variant. Nature 2022, 603, 700–705. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Lu, L.; Peng, Z.; Chen, L.-L.; Meng, X.; Zhang, C.; Ip, J.D.; Chan, W.-M.; Chu, A.W.-H.; Chan, K.-H.; et al. SARS-CoV-2 Omicron variant shows less efficient replication and fusion activity when compared with Delta variant in TMPRSS2-expressed cells. Emerg. Microbes Infect. 2022, 11, 277–283. [Google Scholar] [CrossRef] [PubMed]
- Strobelt, R.; Adler, J.; Paran, N.; Yahalom-Ronen, Y.; Melamed, S.; Politi, B.; Shulman, Z.; Schmiedel, D.; Shaul, Y. Imatinib inhibits SARS-CoV-2 infection by an off-target-mechanism. Sci. Rep. 2022, 12, 5758. [Google Scholar] [CrossRef] [PubMed]
- Turk, V.; Stoka, V.; Vasiljeva, O.; Renko, M.; Sun, T.; Turk, B.; Turk, D. Cysteine cathepsins: From structure, function and regulation to new frontiers. Biochim. Biophys. Acta BBA Proteins Proteom. 2012, 1824, 68–88. [Google Scholar] [CrossRef] [PubMed]
- Mulgaonkar, N.; Wang, H.; Mallawarachchi, S.; Růžek, D.; Martina, B.; Fernando, S. In Silico and in Vitro Evaluation of Imatinib as an Inhibitor for SARS-CoV-2. J. Biomol. Struct. Dyn. 2022, 41, 3052–3061. [Google Scholar] [CrossRef]
- Miller, K.E.; Kim, Y.; Huh, W.-K.; Park, H.-O. Bimolecular Fluorescence Complementation (BiFC) Analysis: Advances and Recent Applications for Genome-Wide Interaction Studies. J. Mol. Biol. 2015, 427, 2039–2055. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.-D.; Chinenov, Y.; Kerppola, T.K. Visualization of Interactions among bZIP and Rel Family Proteins in Living Cells Using Bimolecular Fluorescence Complementation. Mol. Cell 2002, 9, 789–798. [Google Scholar] [CrossRef] [PubMed]
- Yamasoba, D.; Kimura, I.; Nasser, H.; Morioka, Y.; Nao, N.; Ito, J.; Uriu, K.; Tsuda, M.; Zahradnik, J.; Shirakawa, K.; et al. Virological characteristics of the SARS-CoV-2 Omicron BA.2 spike. Cell 2022, 185, 2103–2115.e19. [Google Scholar] [CrossRef] [PubMed]
- Kimura, I.; Yamasoba, D.; Tamura, T.; Nao, N.; Suzuki, T.; Oda, Y.; Mitoma, S.; Ito, J.; Nasser, H.; Zahradnik, J.; et al. Virological characteristics of the SARS-CoV-2 Omicron BA.2 subvariants, including BA.4 and BA.5. Cell 2022, 185, 3992–4007.e16. [Google Scholar] [CrossRef]
- Li, Q.; Wu, J.; Nie, J.; Zhang, L.; Hao, H.; Liu, S.; Zhao, C.; Zhang, Q.; Liu, H.; Nie, L.; et al. The Impact of Mutations in SARS-CoV-2 Spike on Viral Infectivity and Antigenicity. Cell 2020, 182, 1284–1294.e9. [Google Scholar] [CrossRef]
- Toelzer, C.; Gupta, K.; Yadav, S.K.N.; Borucu, U.; Davidson, A.D.; Williamson, M.K.; Shoemark, D.K.; Garzoni, F.; Staufer, O.; Milligan, R.; et al. Free fatty acid binding pocket in the locked structure of SARS-CoV-2 spike protein. Science 2020, 370, eabd3255. [Google Scholar] [CrossRef]
- Carrique, L.; Duyvesteyn, H.M.E.; Malinauskas, T.; Zhao, Y.; Ren, J.; Zhou, D.; Walter, T.S.; Radecke, J.; Huo, J.; Ruza, R.R.; et al. The SARS-CoV-2 Spike harbours a lipid binding pocket which modulates stability of the prefusion trimer. bioRxiv 2020. [Google Scholar] [CrossRef]
- Iwata-Yoshikawa, N.; Okamura, T.; Shimizu, Y.; Hasegawa, H.; Takeda, M.; Nagata, N. TMPRSS2 Contributes to Virus Spread and Immunopathology in the Airways of Murine Models after Coronavirus Infection. J. Virol. 2019, 93, e01815-18. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Chan, J.F.-W.; Liu, H.; Liu, Y.; Chai, Y.; Shi, J.; Shuai, H.; Hou, Y.; Huang, X.; Yuen, T.T.-T.; et al. Spike mutations contributing to the altered entry preference of SARS-CoV-2 Omicron BA.1 and BA.2. Emerg. Microbes Infect. 2022, 11, 2275–2287. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, M.; Tomita, K.; Hirayama, Y.; Inoue, J.-I.; Kawaguchi, Y.; Gohda, J. SARS-CoV-2 Omicron spike H655Y mutation is responsible for enhancement of the endosomal entry pathway and reduction of cell surface entry pathways. bioRxiv 2022. [Google Scholar] [CrossRef]
- Mautner, L.; Hoyos, M.; Dangel, A.; Berger, C.; Ehrhardt, A.; Baiker, A. Replication kinetics and infectivity of SARS-CoV-2 variants of concern in common cell culture models. Virol. J. 2022, 19, 76. [Google Scholar] [CrossRef] [PubMed]
- Peacock, T.P.; Brown, J.C.; Zhou, J.; Thakur, N.; Newman, J.; Kugathasan, R.; Sukhova, K.; Kaforou, M.; Bailey, D.; Barclay, W.S. The SARS-CoV-2 variant, Omicron, shows rapid replication in human primary nasal epithelial cultures and efficiently uses the endosomal route of entry. bioRxiv 2022. [Google Scholar] [CrossRef]
- Lamers, M.M.; Mykytyn, A.Z.; Breugem, T.I.; Groen, N.; Knoops, K.; Schipper, D.; van Acker, R.; Doel, P.B.V.D.; Bestebroer, T.; Koopman, C.D.; et al. SARS-CoV-2 Omicron efficiently infects human airway, but not alveolar epithelium. bioRxiv 2022. [Google Scholar] [CrossRef]
- Wang, S.; Liu, L.; Wang, C.; Wang, Z.; Duan, X.; Chen, G.; Zhou, H.; Shao, H. Establishment of a pseudovirus neutralization assay based on SARS-CoV-2 S protein incorporated into lentiviral particles. Biosaf. Health 2022, 4, 38–44. [Google Scholar] [CrossRef] [PubMed]
- Sholukh, A.M.; Fiore-Gartland, A.; Ford, E.S.; Miner, M.D.; Hou, Y.J.; Tse, L.V.; Kaiser, H.; Zhu, H.; Lu, J.; Madarampalli, B.; et al. Evaluation of Cell-Based and Surrogate SARS-CoV-2 Neutralization Assays. J. Clin. Microbiol. 2021, 59, 527–548. [Google Scholar] [CrossRef]
- Leroy, H.; Han, M.; Woottum, M.; Bracq, L.; Bouchet, J.; Xie, M.; Benichou, S. Virus-Mediated Cell-Cell Fusion. Int. J. Mol. Sci. 2020, 21, 9644. [Google Scholar] [CrossRef]
- Diamond, M.S.; Kanneganti, T.-D. Innate immunity: The first line of defense against SARS-CoV-2. Nat. Immunol. 2022, 23, 165–176. [Google Scholar] [CrossRef]
- Danza, P.; Koo, T.H.; Haddix, M.; Fisher, R.; Traub, E.; Oyong, K.; Balter, S. SARS-CoV-2 Infection and Hospitalization Among Adults Aged ≥18 Years, by Vaccination Status, Before and During SARS-CoV-2 B.1.1.529 (Omicron) Variant Predominance—Los Angeles County, California, November 7, 2021–January 8, 2022. MMWR. Morb. Mortal. Wkly. Rep. 2022, 71, 177–181. [Google Scholar] [CrossRef] [PubMed]
- Nyberg, T.; Ferguson, N.M.; Nash, S.G.; Webster, H.H.; Flaxman, S.; Andrews, N.; Hinsley, W.; Bernal, J.L.; Kall, M.; Bhatt, S.; et al. Comparative analysis of the risks of hospitalisation and death associated with SARS-CoV-2 omicron (B.1.1.529) and delta (B.1.617.2) variants in England: A cohort study. Lancet 2022, 399, 1303–1312. [Google Scholar] [CrossRef] [PubMed]
- Van Goethem, N.; Chung, P.Y.J.; Meurisse, M.; Vandromme, M.; De Mot, L.; Brondeel, R.; Stouten, V.; Klamer, S.; Cuypers, L.; Braeye, T.; et al. Clinical Severity of SARS-CoV-2 Omicron Variant Compared with Delta among Hospitalized COVID-19 Patients in Belgium during Autumn and Winter Season 2021–2022. Viruses 2022, 14, 1297. [Google Scholar] [CrossRef]
- Havers, F.P.; Patel, K.; Whitaker, M.; Milucky, J.; Reingold, A.; Armistead, I.; Meek, J.; Anderson, E.J.; Weigel, A.; Reeg, L.; et al. Laboratory-Confirmed COVID-19–Associated Hospitalizations among Adults During SARS-CoV-2 Omicron BA.2 Variant Predominance—COVID-19–Associated Hospitalization Surveillance Network, 14 States, June 20, 2021–May 31, 2022. MMWR. Morb. Mortal. Wkly. Rep. 2022, 71, 1085–1091. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Strobelt, R.; Broennimann, K.; Adler, J.; Shaul, Y. SARS-CoV-2 Omicron Specific Mutations Affecting Infectivity, Fusogenicity, and Partial TMPRSS2-Independency. Viruses 2023, 15, 1129. https://doi.org/10.3390/v15051129
Strobelt R, Broennimann K, Adler J, Shaul Y. SARS-CoV-2 Omicron Specific Mutations Affecting Infectivity, Fusogenicity, and Partial TMPRSS2-Independency. Viruses. 2023; 15(5):1129. https://doi.org/10.3390/v15051129
Chicago/Turabian StyleStrobelt, Romano, Karin Broennimann, Julia Adler, and Yosef Shaul. 2023. "SARS-CoV-2 Omicron Specific Mutations Affecting Infectivity, Fusogenicity, and Partial TMPRSS2-Independency" Viruses 15, no. 5: 1129. https://doi.org/10.3390/v15051129




