GRP78 Inhibitor YUM70 Suppresses SARS-CoV-2 Viral Entry, Spike Protein Production and Ameliorates Lung Damage
Abstract
:1. Introduction
2. Materials and Methods
2.1. Immunoblot Analysis
2.2. Cell Culture and Drug Treatment
2.3. Expression Vector and Transfection
2.4. Generation of VSV Pseudotyped Viral Particles and Transduction Experiments
2.5. WST-1 Cell Viability Assay
2.6. Virus Propagation
2.7. Plaque Reduction Assay
2.8. Virus Infection, Drug Treatment and Cell Harvest
2.9. Animals and In Vivo Procedures
2.10. Measurement of Magnitude of Pulmonary Damage
2.11. Statistical Analysis
3. Results
3.1. GRP78 Inhibitor YUM70 Blocks Pseudovirus Entry Mediated by Different SARS-CoV-2 Variants Spike Proteins
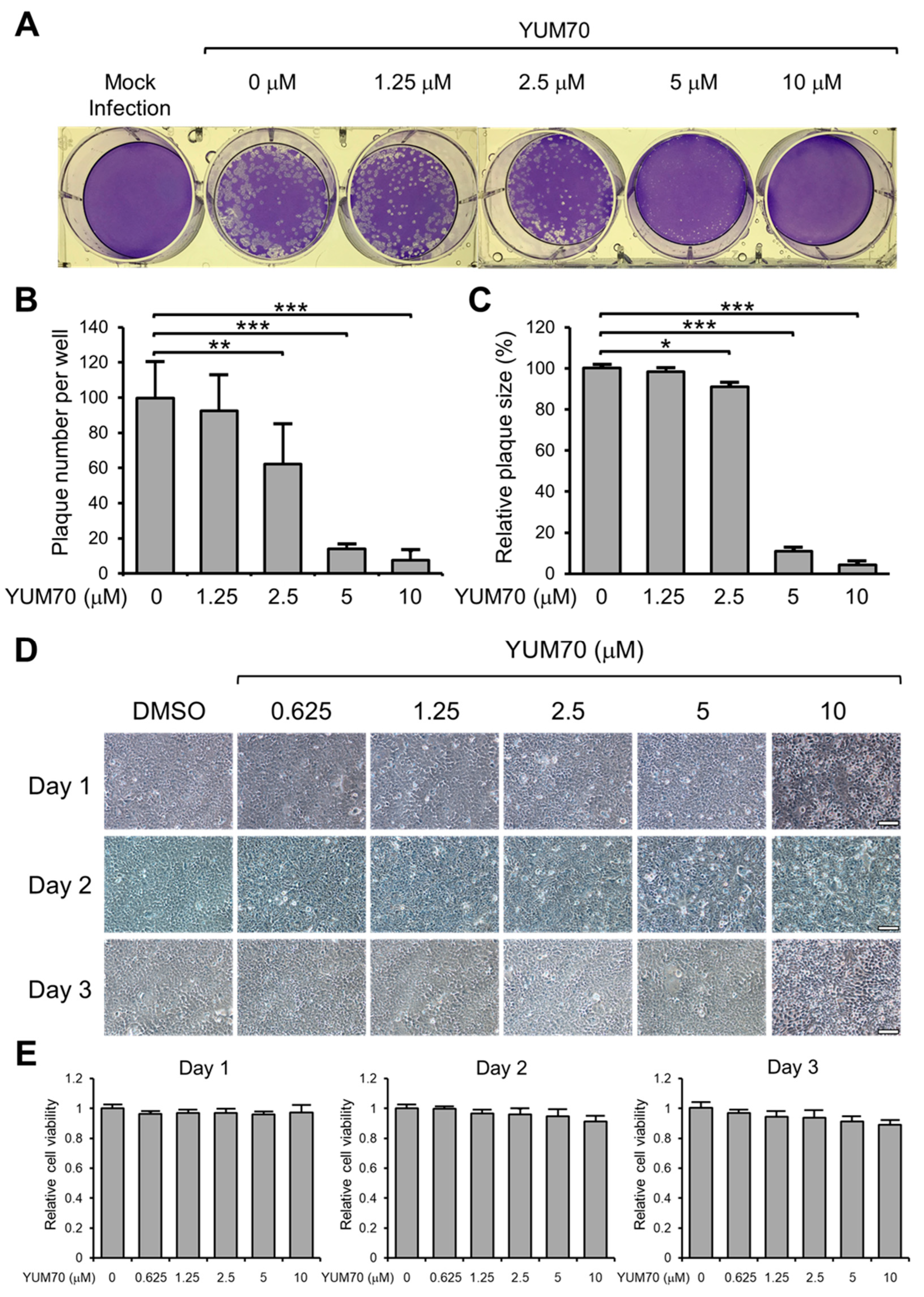
3.2. YUM70 Blocks SARS-CoV-2 Infection In Vitro without Affecting Cell Viability
3.3. YUM70 Reduces Both Spike and N Protein Levels Following SARS-CoV-2 Infection
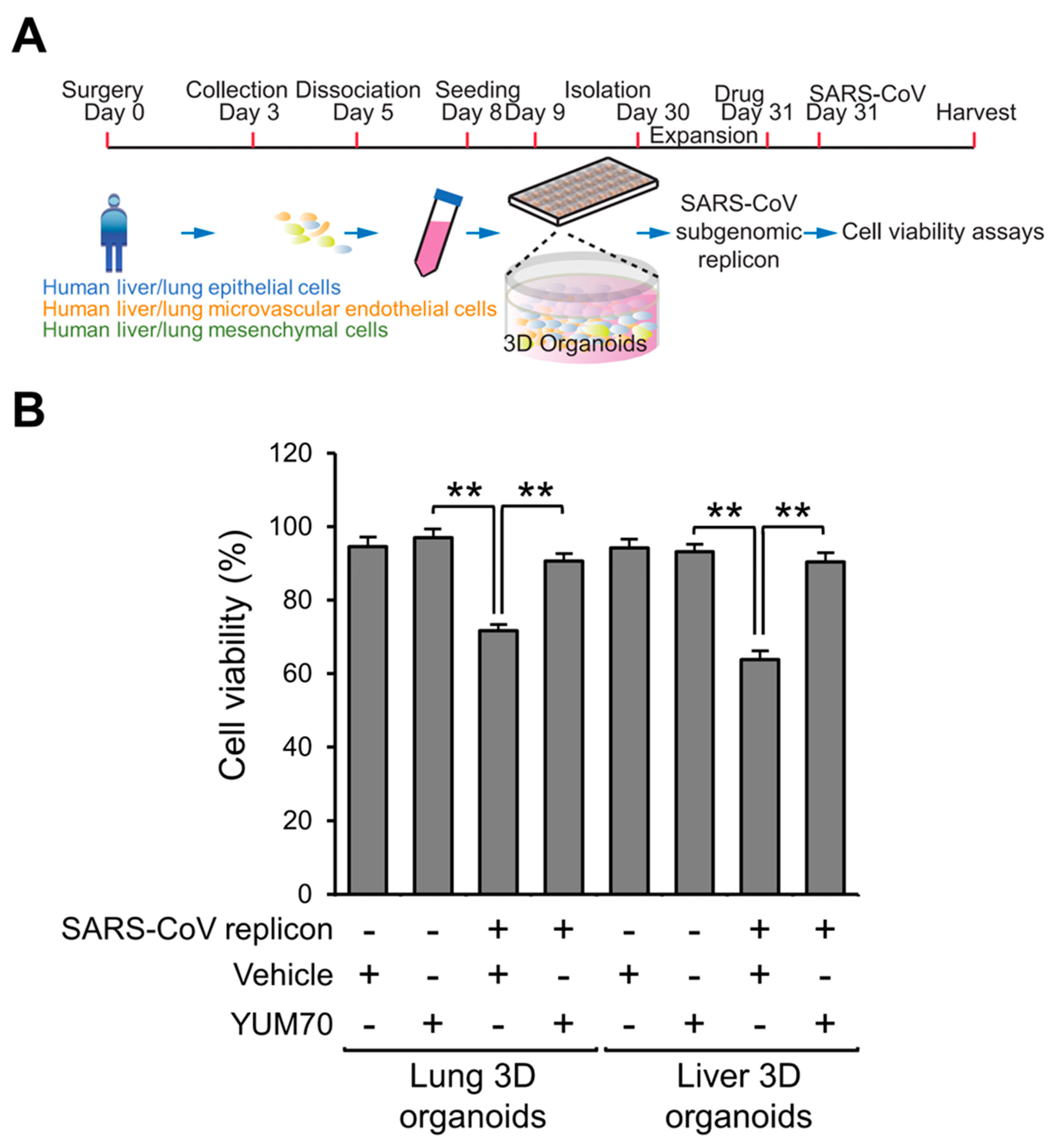
3.4. YUM70 Rescues Cell Viability in Multi-Cellular Human Lung and Liver 3D Organoids Transfected with SARS-CoV-2 Replicon
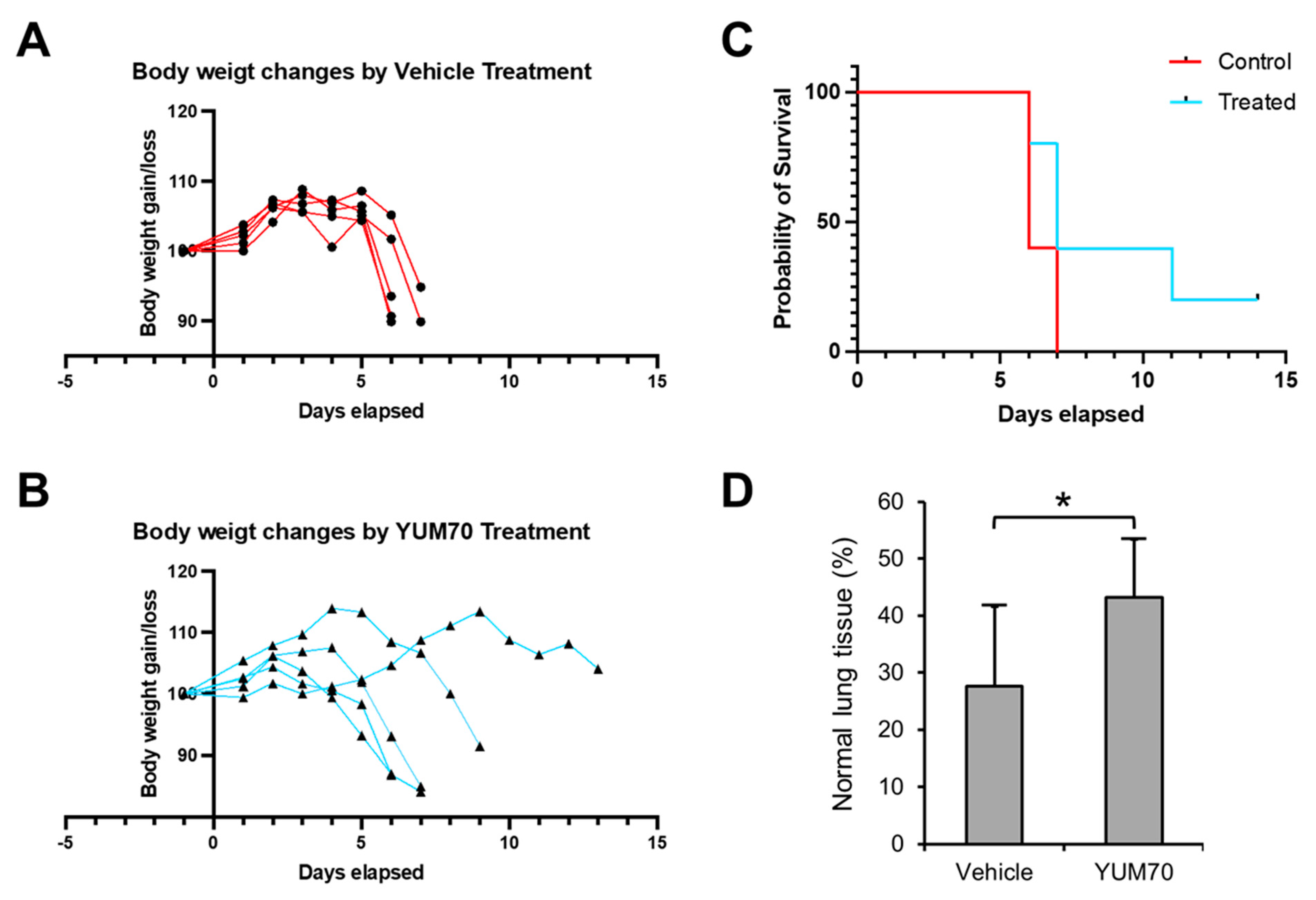
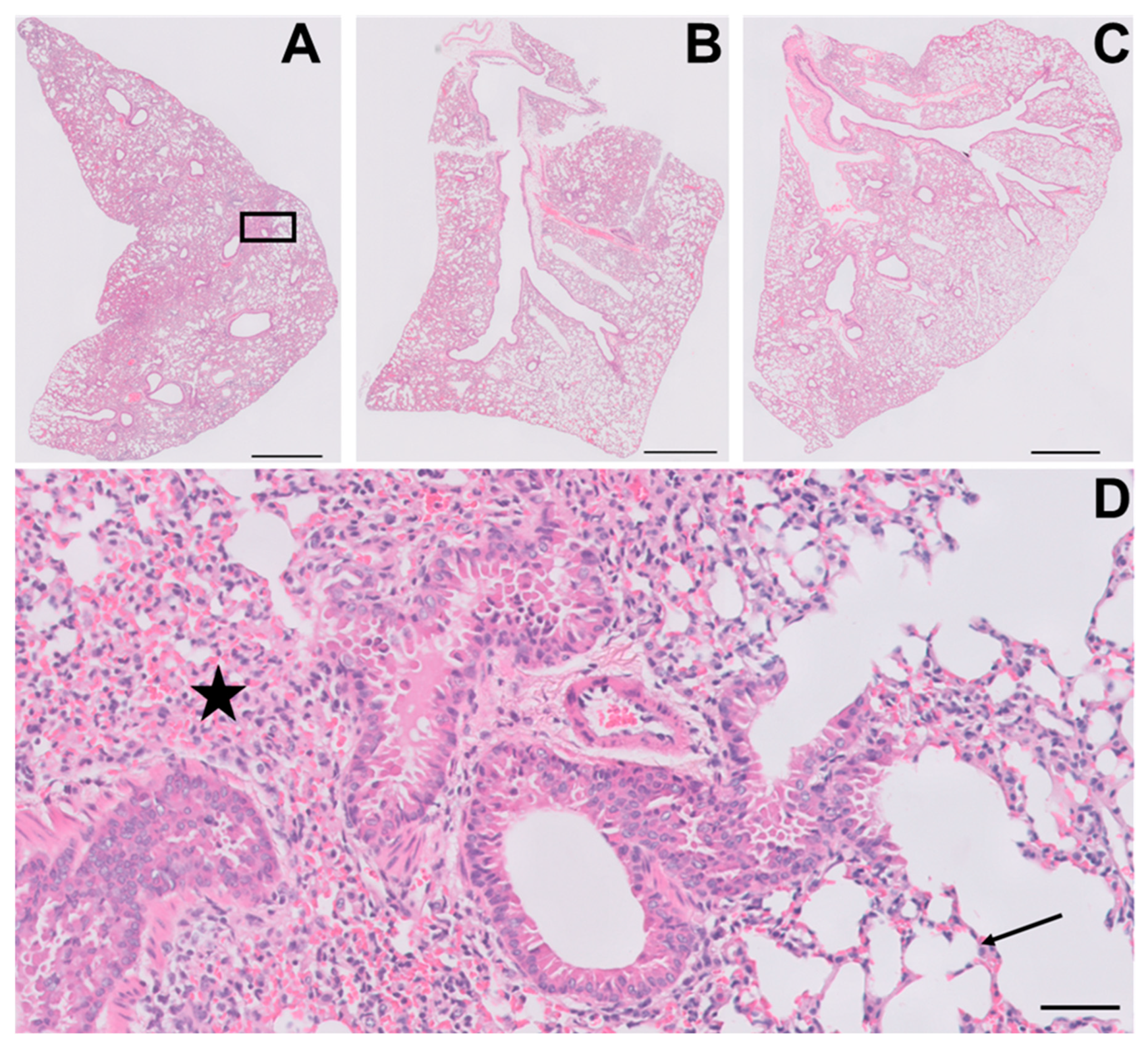
3.5. YUM70 Treated Mice Show Trends of Ameliorated Weight Loss, Morbidity and Lung Damage Following SARS-CoV-2 Infection
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yao, L.; Zhu, K.L.; Jiang, X.L.; Wang, X.J.; Zhan, B.D.; Gao, H.X.; Geng, X.Y.; Duan, L.J.; Dai, E.H.; Ma, M.J. Omicron subvariants escape antibodies elicited by vaccination and BA.2.2 infection. Lancet Infect. Dis. 2022, 22, 1116–1117. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.A.; Song, C.H. Insights Into the Role of Endoplasmic Reticulum Stress in Infectious Diseases. Front. Immunol. 2020, 10, 3147. [Google Scholar] [CrossRef] [PubMed]
- Ha, D.P.; Van Krieken, R.; Carlos, A.J.; Lee, A.S. The stress-inducible molecular chaperone GRP78 as potential therapeutic target for coronavirus infection. J. Infect. 2020, 81, 452–482. [Google Scholar] [CrossRef] [PubMed]
- Kohli, E.; Causse, S.; Baverel, V.; Dubrez, L.; Borges-Bonan, N.; Demidov, O.; Garrido, C. Endoplasmic Reticulum Chaperones in Viral Infection: Therapeutic Perspectives. Microbiol. Mol. Biol. Rev. 2021, 85, e0003521. [Google Scholar] [CrossRef]
- Ni, M.; Lee, A.S. ER chaperones in mammalian development and human diseases. FEBS Lett. 2007, 581, 3641–3651. [Google Scholar] [CrossRef]
- Luo, B.; Lee, A.S. The critical roles of endoplasmic reticulum chaperones and unfolded protein response in tumorigenesis and anticancer therapies. Oncogene 2013, 32, 805–818. [Google Scholar] [CrossRef]
- Lee, A.S. Glucose-regulated proteins in cancer: Molecular mechanisms and therapeutic potential. Nat. Rev. Cancer 2014, 14, 263–276. [Google Scholar] [CrossRef]
- Hetz, C.; Zhang, K.; Kaufman, R.J. Mechanisms, regulation and functions of the unfolded protein response. Nat. Rev. Mol. Cell Biol. 2020, 21, 421–438. [Google Scholar] [CrossRef]
- Casas, C. GRP78 at the Centre of the Stage in Cancer and Neuroprotection. Front. Neurosci. 2017, 11, 177. [Google Scholar] [CrossRef]
- Sato, M.; Yao, V.J.; Arap, W.; Pasqualini, R. GRP78 signaling hub a receptor for targeted tumor therapy. Adv. Genet. 2010, 69, 97–114. [Google Scholar] [CrossRef]
- Ni, M.; Zhang, Y.; Lee, A.S. Beyond the endoplasmic reticulum: Atypical GRP78 in cell viability, signalling and therapeutic targeting. Biochem. J. 2011, 434, 181–188. [Google Scholar] [CrossRef] [PubMed]
- Tsai, Y.L.; Ha, D.P.; Zhao, H.; Carlos, A.J.; Wei, S.; Pun, T.K.; Wu, K.; Zandi, E.; Kelly, K.; Lee, A.S. Endoplasmic reticulum stress activates SRC, relocating chaperones to the cell surface where GRP78/CD109 blocks TGF-β signaling. Proc. Natl. Acad. Sci. USA 2018, 115, E4245–E4254. [Google Scholar] [CrossRef] [PubMed]
- Tseng, C.C.; Stanciauskas, R.; Zhang, P.; Woo, D.; Wu, K.; Kelly, K.; Gill, P.S.; Yu, M.; Pinaud, F.; Lee, A.S. GRP78 regulates CD44v membrane homeostasis and cell spreading in tamoxifen-resistant breast cancer. Life Sci. Alliance 2019, 2, e201900377. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Gronow, M.; Gopal, U.; Austin, R.C.; Pizzo, S.V. Glucose-regulated protein (GRP78) is an important cell surface receptor for viral invasion, cancers, and neurological disorders. IUBMB Life 2021, 73, 843–854. [Google Scholar] [CrossRef] [PubMed]
- Carlos, A.J.; Ha, D.P.; Yeh, D.W.; Van Krieken, R.; Tseng, C.C.; Zhang, P.; Gill, P.; Machida, K.; Lee, A.S. The chaperone GRP78 is a host auxiliary factor for SARS-CoV-2 and GRP78 depleting antibody blocks viral entry and infection. J. Biol. Chem. 2021, 296, 100759. [Google Scholar] [CrossRef]
- Wang, M.; Kaufman, R.J. The impact of the endoplasmic reticulum protein-folding environment on cancer development. Nat. Rev. Cancer 2014, 14, 581–597. [Google Scholar] [CrossRef]
- Clarke, H.J.; Chambers, J.E.; Liniker, E.; Marciniak, S.J. Endoplasmic reticulum stress in malignancy. Cancer Cell 2014, 25, 563–573. [Google Scholar] [CrossRef]
- Chu, H.; Chan, C.M.; Zhang, X.; Wang, Y.; Yuan, S.; Zhou, J.; Au-Yeung, R.K.; Sze, K.H.; Yang, D.; Shuai, H.; et al. Middle East respiratory syndrome coronavirus and bat coronavirus HKU9 both can utilize GRP78 for attachment onto host cells. J. Biol. Chem. 2018, 293, 11709–11726. [Google Scholar] [CrossRef]
- Puzyrenko, A.; Jacobs, E.R.; Sun, Y.; Felix, J.C.; Sheinin, Y.; Ge, L.; Lai, S.; Dai, Q.; Gantner, B.N.; Nanchal, R.; et al. Pneumocytes are distinguished by highly elevated expression of the ER stress biomarker GRP78, a co-receptor for SARS-CoV-2, in COVID-19 autopsies. Cell Stress Chaperones 2021, 26, 859–868. [Google Scholar] [CrossRef]
- Sabirli, R.; Koseler, A.; Goren, T.; Turkcuer, I.; Kurt, O. High GRP78 levels in COVID-19 infection: A case-control study. Life Sci. 2021, 265, 118781. [Google Scholar] [CrossRef]
- Rayner, J.O.; Roberts, R.A.; Kim, J.; Poklepovic, A.; Roberts, J.L.; Booth, L.; Dent, P. AR12 (OSU-03012) suppresses GRP78 expression and inhibits SARS-CoV-2 replication. Biochem. Pharmacol. 2020, 182, 114227. [Google Scholar] [CrossRef] [PubMed]
- Echavarría-Consuegra, L.; Cook, G.M.; Busnadiego, I.; Lefèvre, C.; Keep, S.; Brown, K.; Doyle, N.; Dowgier, G.; Franaszek, K.; Moore, N.A.; et al. Manipulation of the unfolded protein response: A pharmacological strategy against coronavirus infection. PLoS Pathog. 2021, 17, e1009644. [Google Scholar] [CrossRef] [PubMed]
- Sims, A.C.; Mitchell, H.D.; Gralinski, L.E.; Kyle, J.E.; Burnum-Johnson, K.E.; Lam, M.; Fulcher, M.L.; West, A.; Smith, R.D.; Randell, S.H.; et al. Unfolded Protein Response Inhibition Reduces Middle East Respiratory Syndrome Coronavirus-Induced Acute Lung Injury. mBio 2021, 12, e0157221. [Google Scholar] [CrossRef] [PubMed]
- Shin, W.J.; Ha, D.P.; Machida, K.; Lee, A.S. The stress-inducible ER chaperone GRP78/BiP is upregulated during SARS-CoV-2 infection and acts as a pro-viral protein. Nat. Commun. 2022, 13, 6551. [Google Scholar] [CrossRef] [PubMed]
- Samanta, S.; Yang, S.; Debnath, B.; Xue, D.; Kuang, Y.; Ramkumar, K.; Lee, A.S.; Ljungman, M.; Neamati, N. The Hydroxyquinoline Analogue YUM70 Inhibits GRP78 to Induce ER Stress-Mediated Apoptosis in Pancreatic Cancer. Cancer Res. 2021, 81, 1883–1895. [Google Scholar] [CrossRef] [PubMed]
- Ha, D.P.; Huang, B.; Wang, H.; Rangel, D.F.; Van Krieken, R.; Liu, Z.; Samanta, S.; Neamati, N.; Lee, A.S. Targeting GRP78 suppresses oncogenic KRAS protein expression and reduces viability of cancer cells bearing various KRAS mutations. Neoplasia 2022, 33, 100837. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef]
- Berger Rentsch, M.; Zimmer, G. A vesicular stomatitis virus replicon-based bioassay for the rapid and sensitive determination of multi-species type I interferon. PLoS ONE 2011, 6, e25858. [Google Scholar] [CrossRef]
- Wang, W.; Shin, W.J.; Zhang, B.; Choi, Y.; Yoo, J.S.; Zimmerman, M.I.; Frederick, T.E.; Bowman, G.R.; Gross, M.L.; Leung, D.W.; et al. The Cap-Snatching SFTSV Endonuclease Domain Is an Antiviral Target. Cell Rep. 2020, 30, 153–163.e5. [Google Scholar] [CrossRef]
- Angeles-Floriano, T.; Sanjuan-Méndez, A.; Rivera-Torruco, G.; Parra-Ortega, I.; Lopez-Martinez, B.; Martinez-Castro, J.; Marin-Santiago, S.; Alcántara-Hernández, C.; Martínez-Martínez, A.; Márquez-González, H.; et al. Leukocyte surface expression of the endoplasmic reticulum chaperone GRP78 is increased in severe COVID-19. J. Leukoc. Biol. 2023, 113, 1–10. [Google Scholar] [CrossRef]
- Shin, J.; Toyoda, S.; Nishitani, S.; Fukuhara, A.; Kita, S.; Otsuki, M.; Shimomura, I. Possible Involvement of Adipose Tissue in Patients With Older Age, Obesity, and Diabetes With SARS-CoV-2 Infection (COVID-19) via GRP78 (BIP/HSPA5): Significance of Hyperinsulinemia Management in COVID-19. Diabetes 2021, 70, 2745–2755. [Google Scholar] [CrossRef] [PubMed]
- Elfiky, A.A.; Ibrahim, I.M.; Elgohary, A.M. SARS-CoV-2 Delta Variant is Recognized Through GRP78 Host-Cell Surface Receptor, In Silico Perspective. Int. J. Pept. Res. Ther. 2022, 28, 146. [Google Scholar] [CrossRef] [PubMed]
- Elfiky, A.A.; Ibrahim, I.M.; Ibrahim, M.N.; Elshemey, W.M. Host-cell recognition of SARS-CoV-2 spike receptor binding domain from different variants. J. Infect. 2022, 85, 702–769. [Google Scholar] [CrossRef] [PubMed]
- Cerezo, M.; Lehraiki, A.; Millet, A.; Rouaud, F.; Plaisant, M.; Jaune, E.; Botton, T.; Ronco, C.; Abbe, P.; Amdouni, H.; et al. Compounds Triggering ER Stress Exert Anti-Melanoma Effects and Overcome BRAF Inhibitor Resistance. Cancer Cell 2016, 29, 805–819. [Google Scholar] [CrossRef]
- Bakewell, S.J.; Rangel, D.F.; Ha, D.P.; Sethuraman, J.; Crouse, R.; Hadley, E.; Costich, T.L.; Zhou, X.; Nichols, P.; Lee, A.S. Suppression of stress induction of the 78-kilodalton glucose regulated protein (GRP78) in cancer by IT-139, an anti-tumor ruthenium small molecule inhibitor. Oncotarget 2018, 9, 29698–29714. [Google Scholar] [CrossRef]
- Ha, D.P.; Tsai, Y.L.; Lee, A.S. Suppression of ER-stress induction of GRP78 as an anti-neoplastic mechanism of the cardiac glycoside Lanatoside C in pancreatic cancer: Lanatoside C suppresses GRP78 stress induction. Neoplasia 2021, 23, 1213–1226. [Google Scholar] [CrossRef]
- Yamamoto, V.; Wang, B.; Lee, A.S. Suppression of head and neck cancer cell survival and cisplatin resistance by GRP78 small molecule inhibitor YUM70. Front. Oncol. 2023, 12, 1044699. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ha, D.P.; Shin, W.-J.; Hernandez, J.C.; Neamati, N.; Dubeau, L.; Machida, K.; Lee, A.S. GRP78 Inhibitor YUM70 Suppresses SARS-CoV-2 Viral Entry, Spike Protein Production and Ameliorates Lung Damage. Viruses 2023, 15, 1118. https://doi.org/10.3390/v15051118
Ha DP, Shin W-J, Hernandez JC, Neamati N, Dubeau L, Machida K, Lee AS. GRP78 Inhibitor YUM70 Suppresses SARS-CoV-2 Viral Entry, Spike Protein Production and Ameliorates Lung Damage. Viruses. 2023; 15(5):1118. https://doi.org/10.3390/v15051118
Chicago/Turabian StyleHa, Dat P., Woo-Jin Shin, Juan Carlos Hernandez, Nouri Neamati, Louis Dubeau, Keigo Machida, and Amy S. Lee. 2023. "GRP78 Inhibitor YUM70 Suppresses SARS-CoV-2 Viral Entry, Spike Protein Production and Ameliorates Lung Damage" Viruses 15, no. 5: 1118. https://doi.org/10.3390/v15051118




