Porcine Circovirus 2 Increases the Frequency of Transforming Growth Factor-β via the C35, S36 and V39 Amino Acids of the ORF4
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animal, Cell and Virus
2.2. Plasmid Construction and Transfection
2.3. CD4+ T Cell Preparation
2.4. Co-Culture System
2.5. Flow Cytometry
2.6. Extraction of Total RNA and Quantitative RT-PCR
2.7. ELISA
2.8. Immunofluorescence Staining and Confocal Microscopy
2.9. Statistical Analysis
2.10. Ethics Statement
3. Results
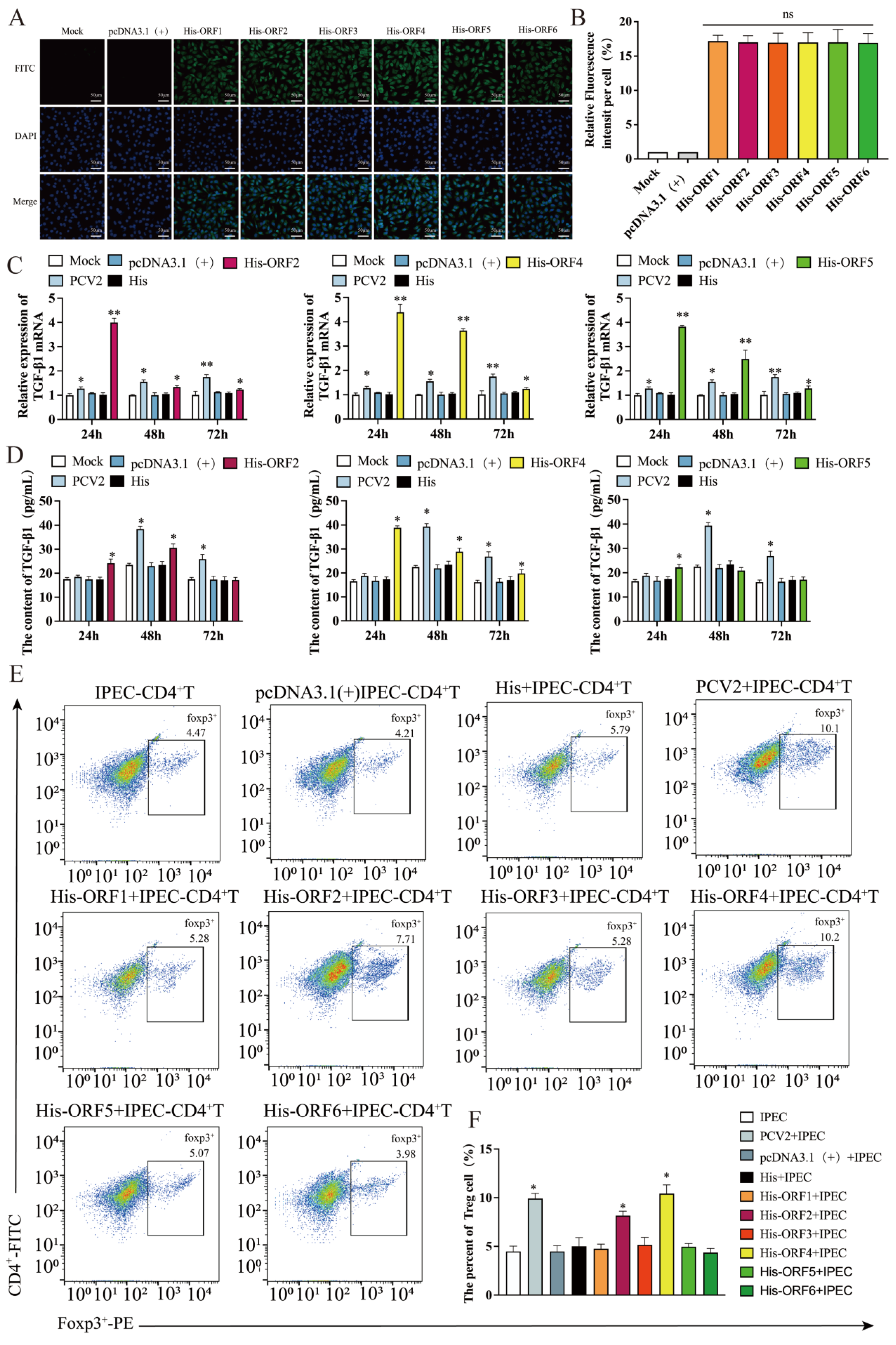
3.1. Proteins Encoded by ORF2, ORF4 and ORF5 Induces the Production of TGF-β in IPEC-J2
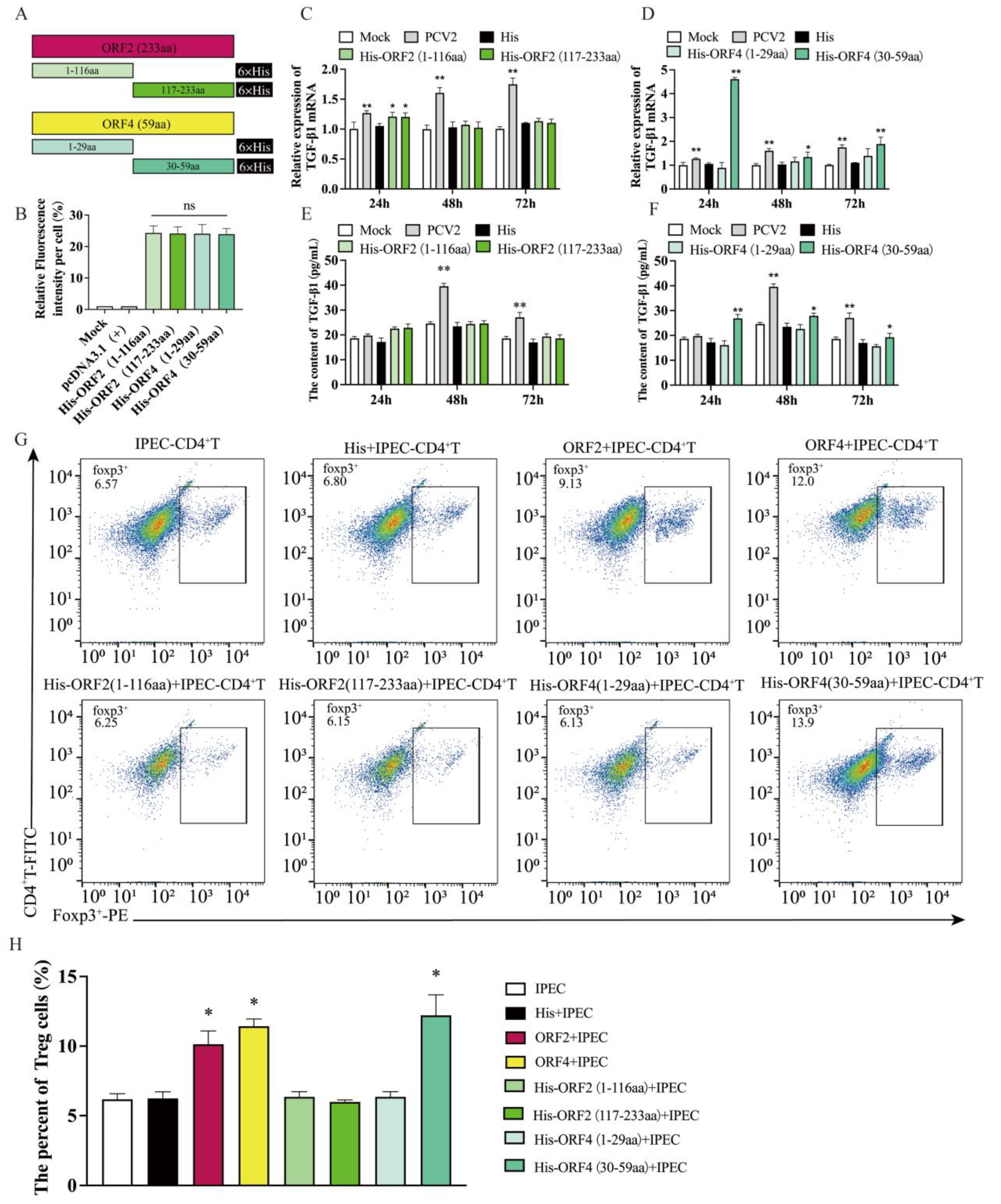
3.2. Overexpression of ORF2 and ORF4 in IPEC-J2 Enhanced the Frequency of Tregs
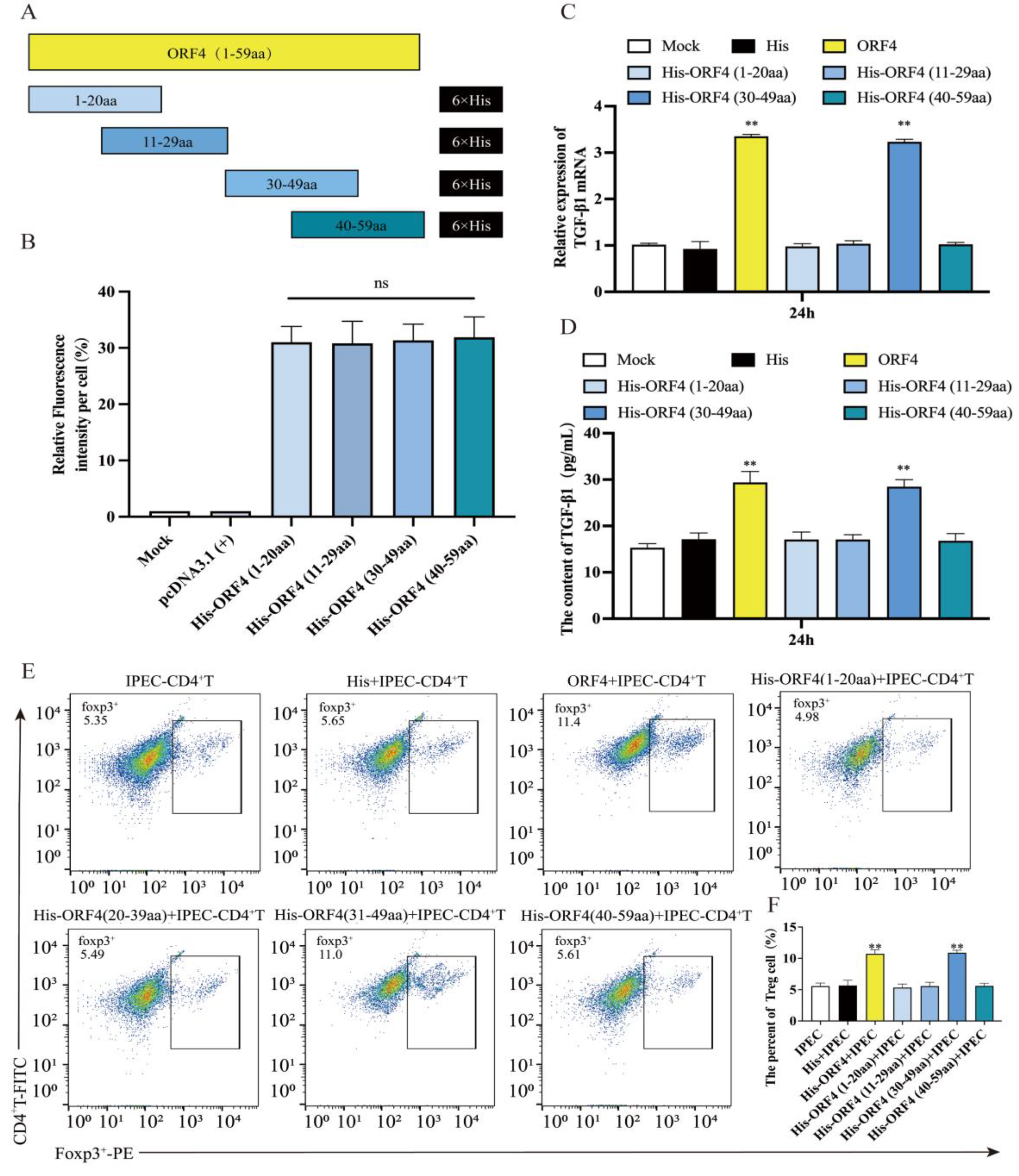
3.3. Overexpression of ORF4 (30–59aa) in IPEC-J2 Enhanced the Frequency of Tregs
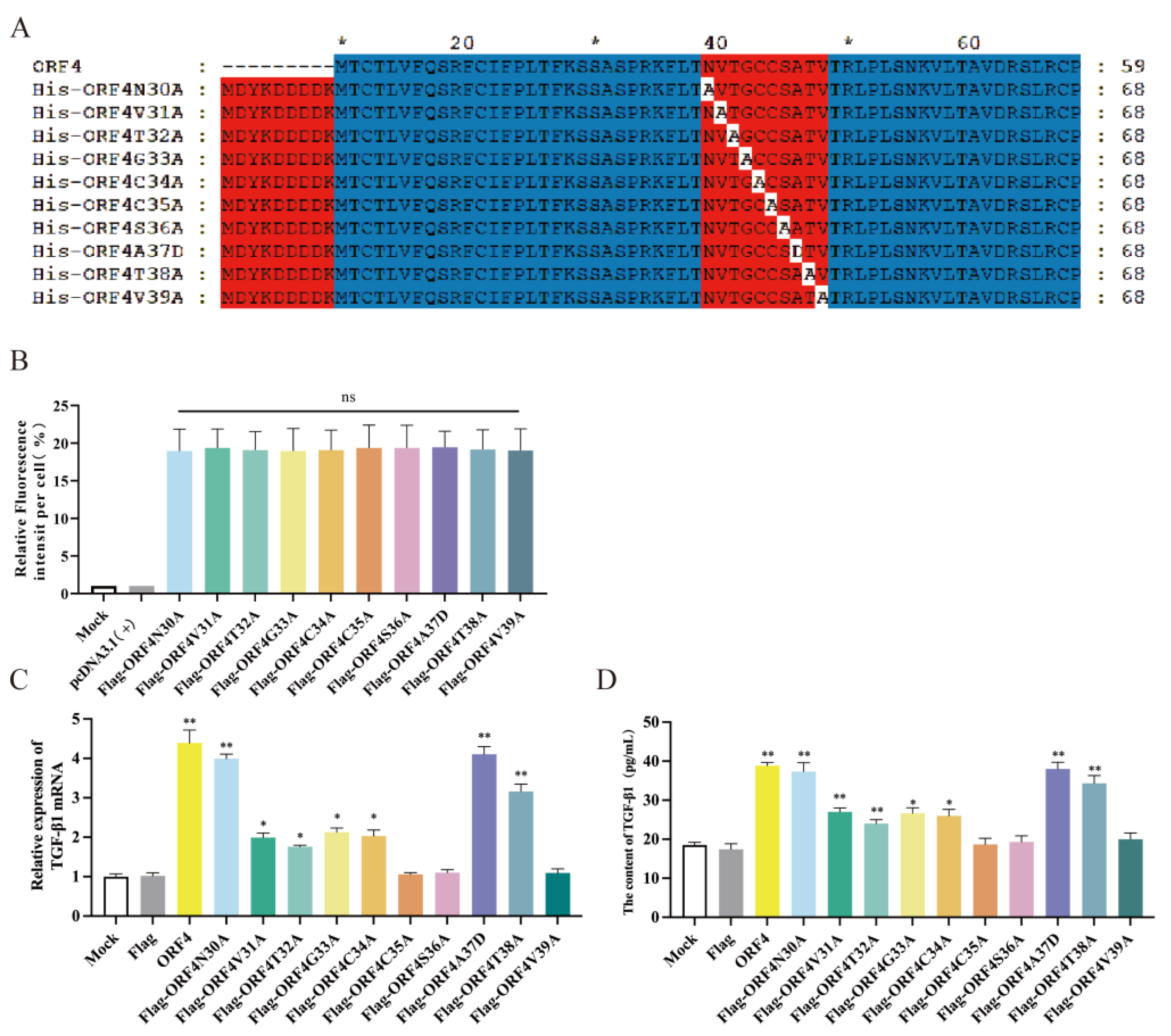
3.4. The 30–39aa of ORF4 Altered the Frequency of Tregs
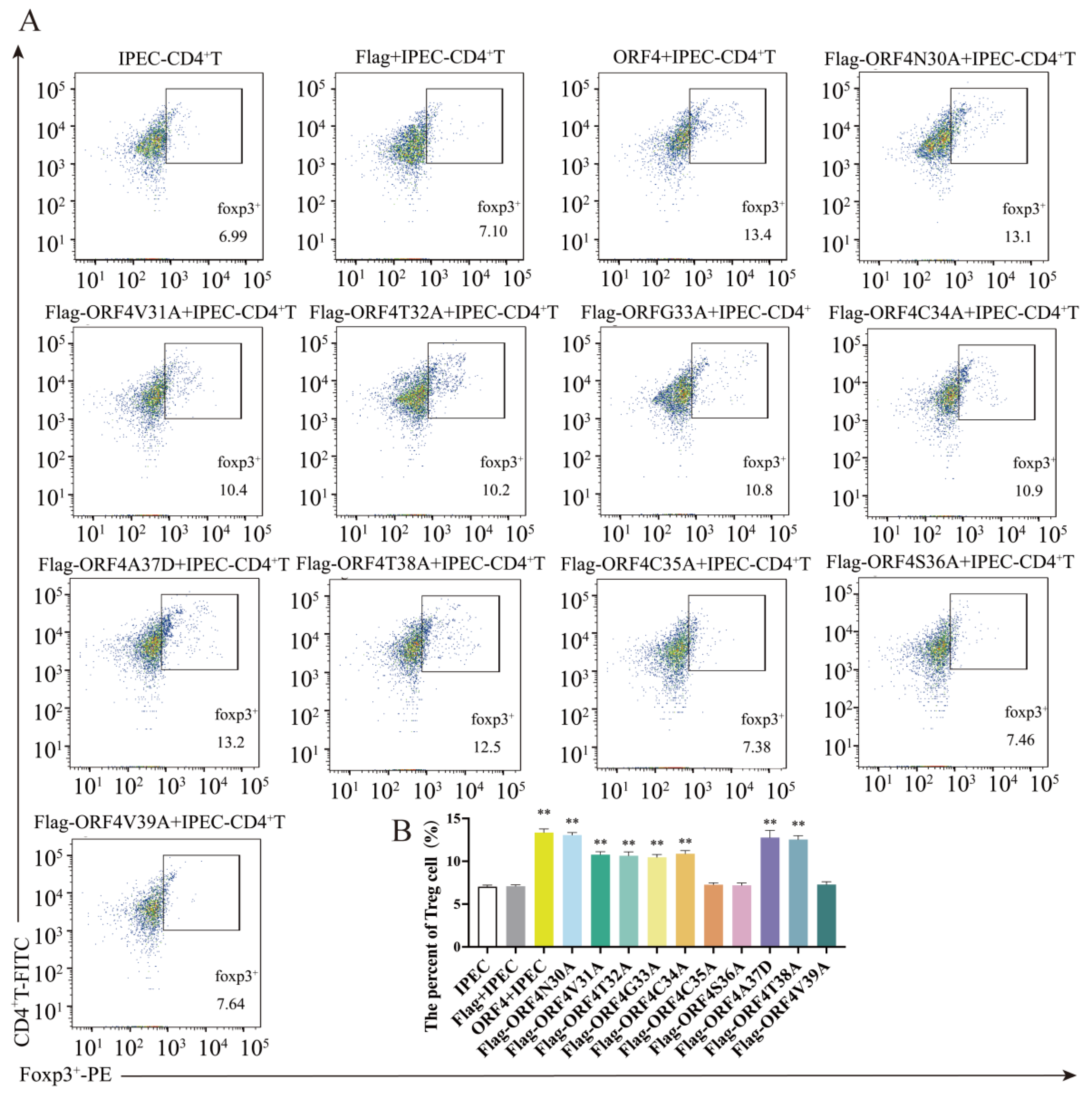
3.5. C35, S36 and V39 Were Key Loci for ORF4-Induced Changes of Tregs Subpopulation
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Allan, G.M.; Mcneilly, F.; Kennedy, S.; Daft, B.; Clarke, E.G.; Ellis, J.A.; Haines, D.M.; Meehan, B.M.; Adair, B.M. Isolation of porcine circovirus-like viruses from pigs with a wasting disease in the USA and Europe. J. Vet. Diagn. Investig. 1998, 10, 3–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Opriessnig, T.; Meng, X.J.; Halbur, P.G. Porcine circovirus type 2 associated disease: Update on current terminology, clinical manifestations, pathogenesis, diagnosis, and intervention strategies. J. Vet. Diagn. Investig. 2007, 19, 591–615. [Google Scholar] [CrossRef]
- Segalés, J. Porcine circovirus type 2 (PCV2) infections: Clinical signs, pathology and laboratory diagnosis. Virus Res. 2012, 164, 10–19. [Google Scholar] [CrossRef] [PubMed]
- Mankertz, A. Molecular interactions of porcine circoviruses type 1 and type 2 with its host. Virus Res. 2012, 164, 54–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheung, A.K. Porcine circovirus: Transcription and DNA replication. Virus Res. 2012, 164, 46–53. [Google Scholar] [CrossRef]
- Ramamoorthy, S.; Meng, X.J. Porcine circoviruses: A minuscule yet mammoth paradox. Anim. Health Res. Rev. 2009, 10, 1–20. [Google Scholar] [CrossRef]
- Zhai, N.; Liu, K.; Li, H.; Liu, Z.; Wang, H.; Korolchuk, V.I.; Carroll, B.; Pan, C.; Gan, F.; Huang, K.; et al. PCV2 replication promoted by oxidative stress is dependent on the regulation of autophagy on apoptosis. Vet. Res. 2019, 50, 19. [Google Scholar] [CrossRef] [Green Version]
- Unterweger, C.; Brunthaler, R.; Auer, A.; Fux, R.; Weissenbacher-Lang, C.; Ladinig, A. Reconsideration of the diagnostic criteria required for PCV2 reproductive disease. Vet. J. 2021, 272, 105660. [Google Scholar] [CrossRef]
- Darwich, L.; Segalés, J.; Domingo, M.; Mateu, E. Changes in CD4 (+), CD8 (+), CD4 (+) CD8 (+), and immunoglobulin M-positive peripheral blood mononuclear cells of postweaning multisystemic wasting syndrome-affected pigs and age-matched uninfected wasted and healthy pigs correlate with lesions and porcine circovirus type 2 load in lymphoid tissues. Clin. Diagn. Lab. Immunol. 2002, 9, 236–242. [Google Scholar]
- Shi, K.C.; Guo, X.; Ge, X.N.; Liu, Q.; Yang, H.-C. Cytokine mRNA expression profiles in peripheral blood mononuclear cells from piglets experimentally co-infected with porcine reproductive and respiratory syndrome virus and porcine circovirus type 2. Vet. Microbiol. 2010, 140, 155–160. [Google Scholar] [CrossRef]
- Sun, N.; Sun, P.; Lv, H.; Sun, Y.; Guo, J.; Wang, Z.; Luo, T.; Wang, S.; Li, H. Matrine displayed antiviral activity in porcine alveolar macrophages co-infected by porcine reproductive and respiratory syndrome virus and porcine circovirus type 2. Sci. Rep. 2016, 6, 24401. [Google Scholar] [CrossRef] [Green Version]
- Han, J.; Zhang, S.; Zhang, Y.; Chen, M.; Lv, Y. Porcine circovirus type 2 increases interleukin-1beta and interleukin-10 production via the MyD88-NF-kappa B signaling pathway in porcine alveolar macrophages in vitro. J. Vet. Sci. 2017, 18, 183–191. [Google Scholar] [CrossRef]
- Kanamori, M.; Nakatsukasa, H.; Okada, M.; Lu, Q.; Yoshimura, A. Induced Regulatory T Cells: Their Development, Stability, and Applications. Trends Immunol. 2016, 37, 803–811. [Google Scholar] [CrossRef]
- Sakaguchi, S.; Miyara, M.; Costantino, C.M.; Hafler, D.A. FOXP3+ regulatory T cells in the human immune system. Nat. Rev. Immunol. 2010, 10, 490–500. [Google Scholar] [CrossRef]
- Savage, P.A.; Klawon, D.E.J.; Miller, C.H. Regulatory T Cell Development. Annu. Rev. Immunol. 2020, 38, 421–453. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Zeng, X.; Yu, S.; Ren, L.; Sun, X.; Long, Y.; Wang, X.; Lu, S.; Song, Y.; Sun, X.-H.; et al. Up-regulated DNA-binding inhibitor Id3 promotes differentiation of regulatory T cell to influence antiviral immunity in chronic hepatitis B virus infection. Life Sci. 2021, 285, 119991. [Google Scholar] [CrossRef] [PubMed]
- Cecere, T.E.; Meng, X.J.; Pelzer, K.; Todd, S.; Beach, N.; Ni, Y.; LeRoith, T. Co-infection of porcine dendritic cells with porcine circovirus type 2a (PCV2a) and genotype II porcine reproductive and respiratory syndrome virus (PRRSV) induces CD4(+)CD25(+)FoxP3(+) T cells in vitro. Vet. Microbiol. 2012, 160, 233–239. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wang, Y.; Han, C.; Li, Q.; Hou, X.; Song, Q.; Zhou, S.; Li, H. TGF-β from the Porcine Intestinal Cell Line IPEC-J2 Induced by Porcine Circovirus 2 Increases the Frequency of Treg Cells via the Activation of ERK (in CD4(+) T Cells) and NF-κB (in IPEC-J2). Viruses 2022, 14, 2466. [Google Scholar] [CrossRef] [PubMed]
- Sun, N.; Zhang, H.; Sun, P.; Khan, A.; Guo, J.; Zheng, X.; Sun, Y.; Fan, K.; Yin, W.; Li, H. Matrine exhibits antiviral activity in a PRRSV/PCV2 co-infected mouse model. Phytomedicine 2020, 77, 153289. [Google Scholar] [CrossRef]
- Kang, L.; Wahaab, A.; Shi, K.; Mustafa, B.E.; Zhang, Y.; Zhang, J.; Li, Z.; Qiu, Y.; Li, B.; Liu, K.; et al. Molecular Epidemic Characteristics and Genetic Evolution of Porcine Circovirus Type 2 (PCV2) in Swine Herds of Shanghai, China. Viruses 2022, 14, 289. [Google Scholar] [CrossRef]
- Marruchella, G.; Valbonetti, L.; Bernabò, N.; Ligios, C. Depletion of follicular dendritic cells in tonsils collected from PMWS-affected pigs. Arch. Virol. 2017, 162, 1281–1287. [Google Scholar] [CrossRef]
- Shi, F.; Li, Q.; Zou, Z.; Wang, Y.; Hou, X.; Zhang, Y.; Song, Q.; Zhou, S.; Li, H. The changes of immune-related molecules within the ileal mucosa of piglets infected with porcine circovirus type 2. J. Vet. Sci. 2020, 21, e78. [Google Scholar] [CrossRef]
- Baró, J.; Segalés, J.; Martínez, J. Porcine circovirus type 2 (PCV2) enteric disease: An independent condition or part of the systemic disease? Vet. Microbiol. 2015, 176, 83–87. [Google Scholar] [CrossRef]
- Lu, J.T.; Xu, A.T.; Shen, J.; Ran, Z.H. Crosstalk between intestinal epithelial cell and adaptive immune cell in intestinal mucosal immunity. J. Gastroenterol. Hepatol. 2017, 32, 975–980. [Google Scholar] [CrossRef]
- Mariani, V.; Palermo, S.; Fiorentini, S.; Lanubile, A.; Giuffra, E. Gene expression study of two widely used pig intestinal epithelial cell lines: IPEC-J2 and IPI-2I. Vet. Immunol. Immunopathol. 2009, 131, 278–284. [Google Scholar] [CrossRef]
- Lan, D.; Tang, C.; Yue, H.; Sun, H.; Cui, L.; Hua, X.; Li, J. Microarray analysis of differentially expressed transcripts in porcine intestinal epithelial cells (IPEC-J2) infected with porcine sapelovirus as a model to study innate immune responses to enteric viruses. Arch. Virol. 2013, 158, 1467–1475. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Alexander, P.B.; Wang, X.F. TGF-β Family Signaling in the Control of Cell Proliferation and Survival. Cold Spring Harb. Perspect. Biol. 2017, 9, a022145. [Google Scholar] [CrossRef] [Green Version]
- Dickinson, M.; Kliszczak, A.E.; Giannoulatou, E.; Peppa, D.; Pellegrino, P.; Williams, I.; Drakesmith, H.; Borrow, P. Dynamics of Transforming Growth Factor (TGF)-β Superfamily Cytokine Induction During HIV-1 Infection Are Distinct from Other Innate Cytokines. Front. Immunol. 2020, 11, 596841. [Google Scholar] [CrossRef]
- Reinhold, D.; Wrenger, S.; Kähne, T.; Ansorge, S. HIV-1 Tat: Immunosuppression via TGF-beta1 induction. Immunol. Today 1999, 20, 384–385. [Google Scholar] [CrossRef]
- Mankertz, A.; Hillenbrand, B. Replication of porcine circovirus type 1 requires two proteins encoded by the viral rep gene. Virology 2001, 279, 429–438. [Google Scholar] [CrossRef] [Green Version]
- Cheung, A.K. Comparative analysis of the transcriptional patterns of pathogenic and nonpathogenic porcine circoviruses. Virology 2003, 310, 41–49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fort, M.; Sibila, M.; Nofrarías, M.; Pérez-Martín, E.; Olvera, A.; Mateu, E.; Segalés, J. Porcine circovirus type 2 (PCV2) Cap and Rep proteins are involved in the development of cell-mediated immunity upon PCV2 infection. Vet. Immunol. Immunopathol. 2010, 137, 226–234. [Google Scholar] [CrossRef] [PubMed]
- Nawagitgul, P.; Morozov, I.; Bolin, S.R.; Harms, P.A.; Sorden, S.D.; Paul, P.S. Open reading frame 2 of porcine circovirus type 2 encodes a major capsid protein. J. Gen. Virol. 2000, 81 Pt 9, 2281–2287. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Cao, J.; Zhou, N.; Jin, Y.; Wu, J.; Zhou, J. Identification and functional analysis of the novel ORF4 protein encoded by porcine circovirus type 2. J. Virol. 2013, 87, 1420–1429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Chen, I.; Du, Q.; Chua, H.; Kwang, J. The ORF3 protein of porcine circovirus type 2 is involved in viral pathogenesis in vivo. J. Virol. 2006, 80, 5065–5073. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Zhu, Y.; Chen, I.; Lau, J.; He, F.; Lau, A.; Wang, Z.; Karuppannan, A.K.; Kwang, J. The ORF3 protein of porcine circovirus type 2 interacts with porcine ubiquitin E3 ligase Pirh2 and facilitates p53 expression in viral infection. J. Virol. 2007, 81, 9560–9567. [Google Scholar] [CrossRef] [Green Version]
- Lv, Q.; Guo, K.; Xu, H.; Wang, T.; Zhang, Y. Identification of Putative ORF5 Protein of Porcine Circovirus Type 2 and Functional Analysis of GFP-Fused ORF5 Protein. PLoS ONE 2015, 10, e0127859. [Google Scholar]
- Franzo, G.; Segalés, J. Porcine circovirus 2 (PCV-2) genotype update and proposal of a new genotyping methodology. PLoS ONE 2018, 13, e0208585. [Google Scholar] [CrossRef] [Green Version]
- Jang, G.; Yoo, H.; Kim, Y.; Yang, K.; Lee, C. Genetic and phylogenetic analysis of porcine circovirus type 2 on Jeju Island, South Korea 2019–2020: Evidence of a novel intergenotypic recombinant. Arch. Virol. 2021, 166, 1093–1102. [Google Scholar] [CrossRef]
- Rajkhowa, T.K.; Lalnunthanga, P.; Rao, P.L.; Subbiah, M.; Lalrohlua, B. Emergence of porcine circovirus 2g (PCV2g) and evidence for recombination between genotypes 2g, 2b and 2d among field isolates from non-vaccinated pigs in Mizoram, India. Infect. Genet. Evol. 2021, 90, 104775. [Google Scholar] [CrossRef]
| Genes | Primer Sequence (5′-3′) |
|---|---|
| ORF1 | Forward: CTAGCGTTTAAACTTAAGCTTATGCCCAGTAAGAAGAATGGAAGA Reverse: ATGGTGGCGACCGGTGGATCCTCAATGATGATGATGATGATGGTAA |
| ORF2 | Forward: CTAGCGTTTAAACTTAAGCTTATGACGTATCCAAGGAGGCGT Reverse: ATGGTGGCGACCGGTGGATCCTCAATGATGATGATGATGATGCTTA |
| ORF3 | Forward: CTAGCGTTTAAACTTAAGCTTATGGTAACCATCCCACCACTTG Reverse: ATGGTGGCGACCGGTGGATCCTCAATGATGATGATGATGATGTGTCC |
| ORF4 | Forward: CTAGCGTTTAAACTTAAGCTTATGCCCAGTAAGAAGAATGGAAGA Reverse: ATGGTGGCGACCGGTGGATCCCTAATGATGATGATGATGATGCAAAC |
| ORF5 | Forward: CTAGCGTTTAAACTTAAGCTTATGGTTTTTATTATTCATTTAGGGTTTAAC Reverse: ATGGTGGCGACCGGTGGATCCCTAATGATGATGATGATGATGCAAAC |
| ORF6 | Forward: CTAGCGTTTAAACTTAAGCTTATGGCATCTTCAACACCCGC Reverse: ATGGTGGCGACCGGTGGATCCTCAATGATGATGATGATGATGTGTCC |
| ORF2 1–116A | Forward: CACACTGGACTAGTGGATCCATGCATCATCATCATCATCATACGTATCCAAGGAGG Reverse: CAGCGGTTTAAACTTAAGCTTTCACCTGTCACCCTGGGTGAGT |
| ORF2 117–233A | Forward: CACACTGGACTAGTGGATCCATGCATCATCATCATCATCATAGGAGTGGGCTCCAC Reverse: CAGCGGTTTAAACTTAAGCTTTCAAGGGTTAAGTGGGGGGT |
| ORF4 1–29A | Forward: CCACACTGGACTAGTGGATCCATGCATCATCATCATCATCATACCTGCACCCTGGTG Reverse: CAGCGGTTTAAACTTAAGCTTTCAGTTGGTCAGGAATTTGCG |
| ORF4 30–59A | Forward: CCACACTGGACTAGTGGATCCATGCATCATCATCATCATCATGTGACCGGCTGC Reverse: CAGCGGTTTAAACTTAAGCTTTCATGGGCACCTCAGGGAG |
| ORF4 1–20A | Forward: TAGCGTTTAAACTTAAGCTTATGCATCATCATCATCATCATACCTGCACCCTGGTG Reverse: CCACACTGGACTAGTGGATCCTCAGCTCTTGAAGGTCAGGGG |
| ORF4 11–29A | Forward: CTAGCGTTTAAACTTAAGCTTATGCATCATCATCATCATCATTGCATCTTCCC Reverse: CCACACTGGACTAGTGGATCCTCAGTTGGTCAGGAATTTGCG |
| ORF4 30–49A | Forward: CTAGCGTTTAAACTTAAGCTTATGCATCATCATCATCATCATGTGACCGGCTGC Reverse: CCACACTGGACTAGTGGATCCTCATGGGCACCTCAGGGAG |
| ORF4 40–59A | Forward: CTAGCGTTTAAACTTAAGCTTATGCATCATCATCATCATCATGTGACCGGCTG Reverse: CCACACTGGACTAGTGGATCCTCACAGCACTTTGTTGCTCAGG |
| ORF4N30A | Forward: ATGGATTACAAGGACGACGATGACAAGATGACCTGCAC Reverse: GCCGGTGGCGTTGGT Forward: GGTCACGGCGGTCAG Reverse: TGATTCTCATCAAGCAGGTCTCC |
| ORF4V31A | Forward: ATGGATTACAAGGACGACGATGACAAGATGACCTGCAC Reverse: GCCGGTGGCGTTGGT Forward: ACCAACGCCACCGGC Reverse: TGATTCTCATCAAGCAGGTCTCC |
| ORF4T32A | Forward: ATGGATTACAAGGACGACGATGACAAGATGACCTGCAC Reverse: GCAGCCGGCCACGTT Forward: AACGTGGCCGGCTGC Reverse: TGATTCTCATCAAGCAGGTCTCC |
| ORF4G33A | Forward: ATGGATTACAAGGACGACGATGACAAGATGACCTGCAC Reverse: GCAGCAGGCGGTCAC Forward: GTGACCGCCTGCTGC Reverse: TGATTCTCATCAAGCAGGTCTCC |
| ORF4V34A | Forward: ATGGATTACAAGGACGACGATGACAAGATGACCTGCAC Reverse: GGAGCAGGCGCCGGT Forward: ACCGGCGCCTGCTCC Reverse: TGATTCTCATCAAGCAGGTCTCC |
| ORF4V35A | Forward: ATGGATTACAAGGACGACGATGACAAGATGACCTGCAC Reverse: GGCGGAGGCGCAGCC Forward: GGCTGCGCCTCCGCC Reverse: TGATTCTCATCAAGCAGGTCTCC |
| ORF4S36A | Forward: ATGGATTACAAGGACGACGATGACAAGATGACCTGCAC Reverse: GGTGGCGGCGCAGCA Forward: TGCTGCGCCGCCACC Reverse: TGATTCTCATCAAGCAGGTCTCC |
| ORF4A37D | Forward: ATGGATTACAAGGACGACGATGACAAGATGACCTGCAC Reverse: CACGGTGTCGGAGCA Forward: TGCTCCGACACCGTG Reverse: TGATTCTCATCAAGCAGGTCTCC |
| ORF4T38A | Forward: ATGGATTACAAGGACGACGATGACAAGATGACCTGCAC Reverse: GGTCACGGCGGCGGA Forward: TCCGCCgccGTGACC Reverse: TGATTCTCATCAAGCAGGTCTCC |
| ORF4V39A | Forward: ATGGATTACAAGGACGACGATGACAAGATGACCTGCAC Reverse: CCTGGTGGCGGTGGC Forward: GCCACCGCCACCAGG Reverse: TGATTCTCATCAAGCAGGTCTCC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, C.; Xu, W.; Wang, J.; Hou, X.; Zhou, S.; Song, Q.; Liu, X.; Li, H. Porcine Circovirus 2 Increases the Frequency of Transforming Growth Factor-β via the C35, S36 and V39 Amino Acids of the ORF4. Viruses 2023, 15, 1602. https://doi.org/10.3390/v15071602
Han C, Xu W, Wang J, Hou X, Zhou S, Song Q, Liu X, Li H. Porcine Circovirus 2 Increases the Frequency of Transforming Growth Factor-β via the C35, S36 and V39 Amino Acids of the ORF4. Viruses. 2023; 15(7):1602. https://doi.org/10.3390/v15071602
Chicago/Turabian StyleHan, Cheng, Weicheng Xu, Jianfang Wang, Xiaolin Hou, Shuanghai Zhou, Qinye Song, Xuewei Liu, and Huanrong Li. 2023. "Porcine Circovirus 2 Increases the Frequency of Transforming Growth Factor-β via the C35, S36 and V39 Amino Acids of the ORF4" Viruses 15, no. 7: 1602. https://doi.org/10.3390/v15071602






