Multiplex Serology for Sensitive and Specific Flavivirus IgG Detection: Addition of Envelope Protein Domain III to NS1 Increases Sensitivity for Tick-Borne Encephalitis Virus IgG Detection
Abstract
:1. Introduction
2. Materials and Methods
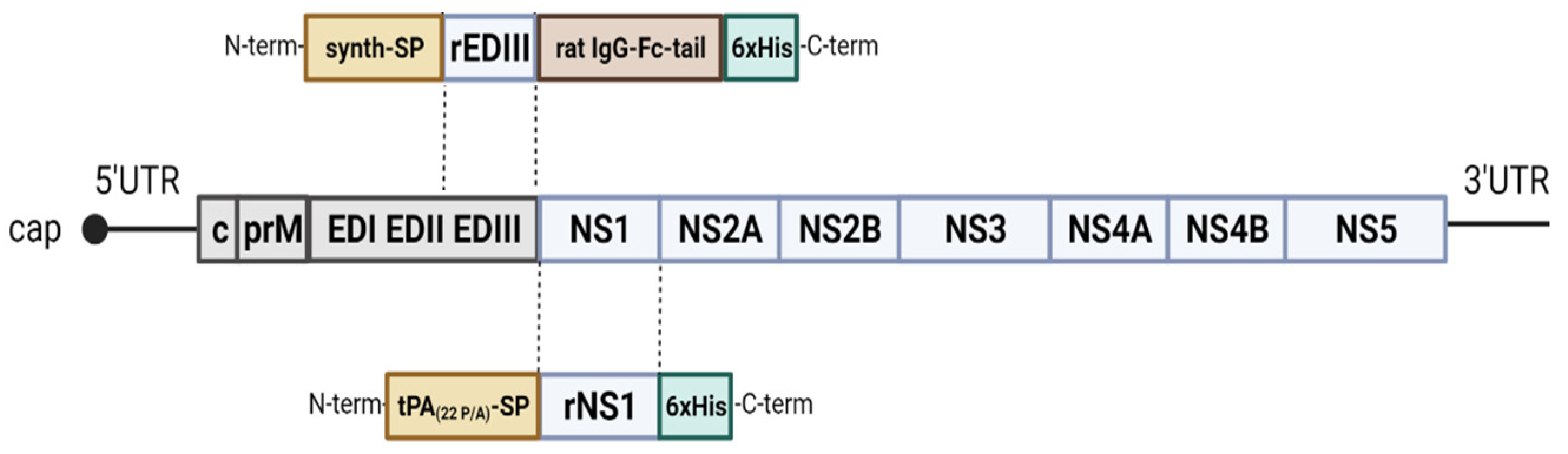
2.1. Cloning and Expression
2.2. Purification of the Recombinant Proteins
2.3. Serum Samples for Multiplex Protein Microarray Validation
2.4. Multiplex Protein Microarray
2.5. Data Analysis
2.6. Plaque Reduction Neutralization Test (PRNT)
2.7. Statistical Analysis
3. Results
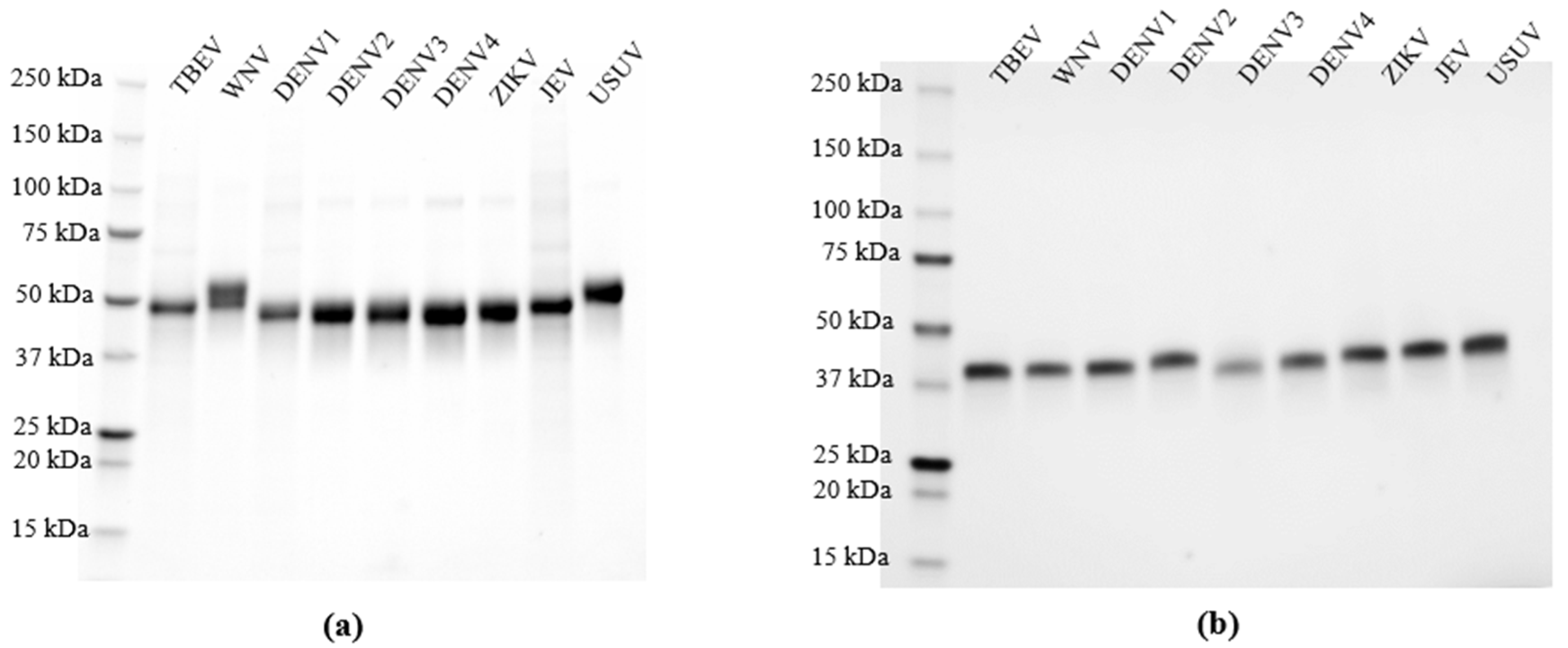
3.1. Expression and Validation of Recombinant NS1 for Specific Orthoflavivirus IgG Detection Using Multiplex Protein Microarray
3.2. WNV and TBEV Plaque Reduction Neutralization Test
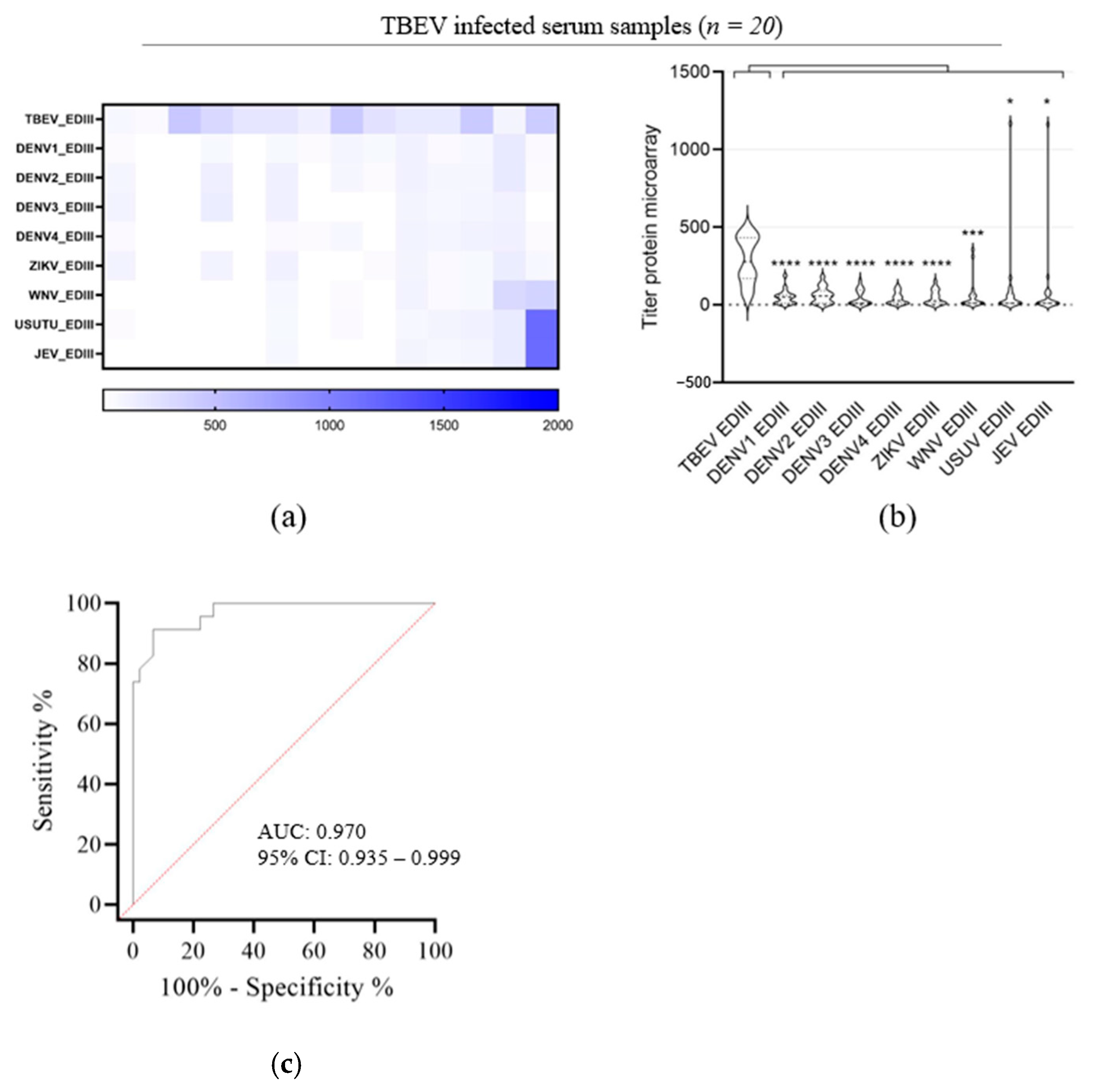
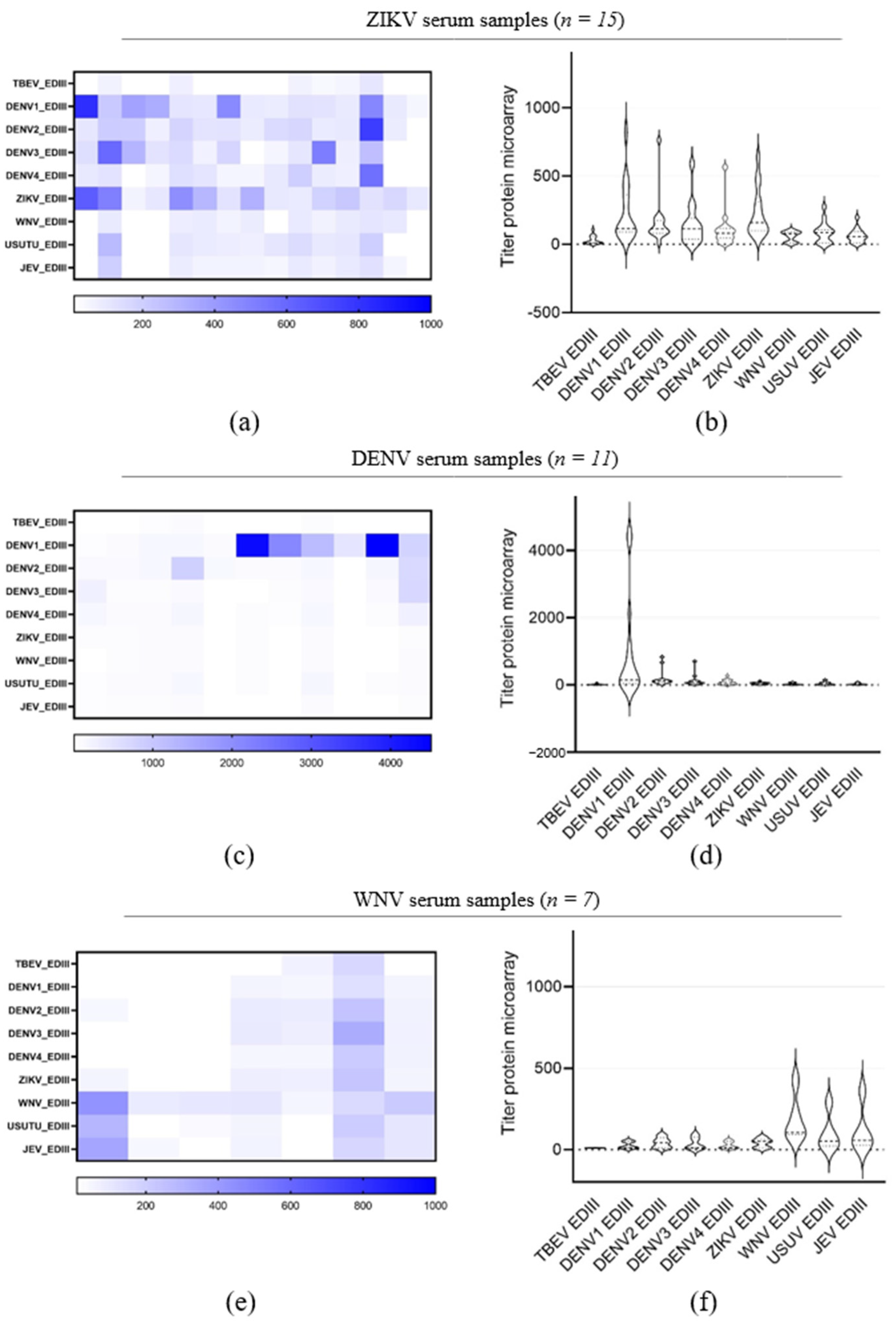
3.3. Evaluation of TBEV EDIII as Antigen for TBEV IgG Detection
3.4. Performance Assessment of rNS1/rEDIII Microarray for TBEV IgG Detection in High-Risk Cohort of Forestry Workers
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bogovic, P.; Strle, F. Tick-borne encephalitis: A review of epidemiology, clinical characteristics, and management. World J. Clin. Cases 2015, 3, 430–441. [Google Scholar] [CrossRef] [PubMed]
- Beauté, J.; Spiteri, G.; Warns-Petit, E.; Zeller, H. Tick-borne encephalitis in Europe, 2012 to 2016. Euro Surveill. Bull. Eur. Sur Mal. Transm. Eur. Commun. Dis. Bull. 2018, 23, 1800201. [Google Scholar] [CrossRef] [PubMed]
- Dekker, M.; Laverman, G.D.; de Vries, A.; Reimerink, J.; Geeraedts, F. Emergence of tick-borne encephalitis (TBE) in the Netherlands. Ticks Tick-Borne Dis. 2019, 10, 176–179. [Google Scholar] [CrossRef] [PubMed]
- Reimerink, J.; Sprong, H.; Harms, M.; Reusken, C.B.E.M. Netherlands, The TBE Book, 6th ed.; Global Health Press: Singapore, 2023; Available online: https://tbenews.com/The_TBE_Book_6th_Edition.pdf (accessed on 24 October 2023).
- Dumpis, U.; Crook, D.; Oksi, J. Tick-borne encephalitis. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 1999, 28, 882–890. [Google Scholar] [CrossRef] [PubMed]
- Ličková, M.; Havlíková, S.F.; Sláviková, M.; Slovák, M.; Drexler, J.F.; Klempa, B. Dermacentor reticulatus is a vector of tick-borne encephalitis virus. Ticks Tick-Borne Dis. 2020, 11, 101414. [Google Scholar] [CrossRef]
- Chitimia-Dobler, L.; Lemhöfer, G.; Król, N.; Bestehorn, M.; Dobler, G.; Pfeffer, M. Repeated isolation of tick-borne encephalitis virus from adult Dermacentor reticulatus ticks in an endemic area in Germany. Parasites Vectors 2019, 12, 90. [Google Scholar] [CrossRef]
- Lindquist, L.; Vapalahti, O. Tick-borne encephalitis. Lancet 2008, 371, 1861–1871. [Google Scholar] [CrossRef]
- Kaiser, R. Tick-Borne Encephalitis. Infect. Dis. Clin. North Am. 2008, 22, 561–575. [Google Scholar] [CrossRef]
- Dobler, G.; Kaier, K.; Hehn, P.; Böhmer, M.; Kreusch, T.; Borde, J. Tick-borne encephalitis virus vaccination breakthrough infections in Germany: A retrospective analysis from 2001 to 2018. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2020, 26, 1090.e7–1090.e13. [Google Scholar] [CrossRef]
- Holzmann, H. Diagnosis of tick-borne encephalitis. Vaccine 2003, 21 (Suppl. S1), S36–S40. [Google Scholar] [CrossRef]
- Kovalev, S.Y.; Mukhacheva, T.A. Reconsidering the classification of tick-borne encephalitis virus within the Siberian subtype gives new insights into its evolutionary history. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2017, 55, 159–165. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Shang, G.; Lu, S.; Yang, J.; Xu, J. A new subtype of eastern tick-borne encephalitis virus discovered in Qinghai-Tibet Plateau, China. Emerg. Microbes Infect. 2018, 7, 74. [Google Scholar] [CrossRef] [PubMed]
- Chambers, T.J.; Hahn, C.S.; Galler, R.; Rice, C.M. Flavivirus genome organization, expression, and replication. Annu. Rev. Microbiol. 1990, 44, 649–688. [Google Scholar] [CrossRef] [PubMed]
- Winkler, G.; Randolph, V.B.; Cleaves, G.R.; Ryan, T.E.; Stollar, V. Evidence that the mature form of the flavivirus nonstructural protein NS1 is a dimer. Virology 1988, 162, 187–196. [Google Scholar] [CrossRef] [PubMed]
- Pryor, M.J.; Wright, P.J. Glycosylation Mutants of Dengue Virus NS1 Protein. J. Gen. Virol. 1994, 75 Pt 5, 1183–1187. [Google Scholar] [CrossRef] [PubMed]
- Alcon, S.; Talarmin, A.; Debruyne, M.; Falconar, A.; Deubel, V.; Flamand, M. Enzyme-linked immunosorbent assay specific to dengue virus type 1 nonstructural protein NS1 reveals circulation of the antigen in the blood during the acute phase of disease in patients experiencing primary or secondary infections. J. Clin. Microbiol. 2002, 40, 376–381. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.-X.; Li, X.-F.; Deng, Y.-Q.; Guo, Y.-H.; Hao, W.; Che, X.-Y.; Qin, C.-F.; Fu, N. Development of a double antibody sandwich ELISA for West Nile virus detection using monoclonal antibodies against non-structural protein 1. PLoS ONE 2014, 9, e108623. [Google Scholar] [CrossRef]
- Kumar, J.S.; Parida, M.; Rao, P.L. Monoclonal antibody-based antigen capture immunoassay for detection of circulating non-structural protein NS1: Implications for early diagnosis of japanese encephalitis virus infection. J. Med. Virol. 2011, 83, 1063–1070. [Google Scholar] [CrossRef]
- Mora-Cárdenas, E.; Aloise, C.; Faoro, V.; Gašper, N.K.; Korva, M.; Caracciolo, I.; D’agaro, P.; Avšič-Županc, T.; Marcello, A. Comparative specificity and sensitivity of NS1-based serological assays for the detection of flavivirus immune response. PLoS Neglected Trop. Dis. 2020, 14, e0008039. [Google Scholar] [CrossRef]
- Rey, F.A.; Heinz, F.X.; Mandl, C.; Kunz, C.; Harrison, S.C. The envelope glycoprotein from tick-borne encephalitis virus at 2 Å resolution. Nature 1995, 375, 291–298. [Google Scholar] [CrossRef]
- Modis, Y.; Ogata, S.; Clements, D.; Harrison, S.C. A ligand-binding pocket in the dengue virus envelope glycoprotein. Proc. Natl. Acad. Sci. USA 2003, 100, 6986–6991. [Google Scholar] [CrossRef]
- Modis, Y.; Ogata, S.; Clements, D.; Harrison, S.C. Variable surface epitopes in the crystal structure of dengue virus type 3 envelope glycoprotein. J. Virol. 2005, 79, 1223–1231. [Google Scholar] [CrossRef]
- Nybakken, G.E.; Nelson, C.A.; Chen, B.R.; Diamond, M.S.; Fremont, D.H. Crystal structure of the West Nile virus envelope glycoprotein. J. Virol. 2006, 80, 11467–11474. [Google Scholar] [CrossRef] [PubMed]
- Wahala, W.; Kraus, A.A.; Haymore, L.B.; Accavitti-Loper, M.A.; de Silva, A.M. Dengue virus neutralization by human immune sera: Role of envelope protein domain III-reactive antibody. Virology 2009, 392, 103–113. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.-W.; Liu, S.-J.; Li, Y.-S.; Liu, H.-H.; Tsai, J.-P.; Chiang, C.-Y.; Chen, M.-Y.; Hwang, C.-S.; Huang, C.-C.; Hu, H.-M.; et al. A consensus envelope protein domain III can induce neutralizing antibody responses against serotype 2 of dengue virus in non-human primates. Arch. Virol. 2013, 158, 1523–1531. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Qi, J.; Peng, R.; Dai, L.; Gould, E.A.; Gao, G.F.; Tien, P. Molecular Basis of a Protective/Neutralizing Monoclonal Antibody Targeting Envelope Proteins of both Tick-Borne Encephalitis Virus and Louping Ill Virus. J. Virol. 2019, 93, e02132-18. [Google Scholar] [CrossRef] [PubMed]
- Tai, W.; He, L.; Wang, Y.; Sun, S.; Zhao, G.; Luo, C.; Li, P.; Zhao, H.; Fremont, D.H.; Li, F.; et al. Critical neutralizing fragment of Zika virus EDIII elicits cross-neutralization and protection against divergent Zika viruses. Emerg. Microbes Infect. 2018, 7, 7. [Google Scholar] [CrossRef]
- Beasley, D.W.C.; Barrett, A.D.T. Identification of neutralizing epitopes within structural domain III of the West Nile virus envelope protein. J. Virol. 2002, 76, 13097–13100. [Google Scholar] [CrossRef]
- Agudelo, M.; Palus, M.; Keeffe, J.R.; Bianchini, F.; Svoboda, P.; Salát, J.; Peace, A.; Gazumyan, A.; Cipolla, M.; Kapoor, T.; et al. Broad and potent neutralizing human antibodies to tick-borne flaviviruses protect mice from disease. J. Exp. Med. 2021, 218, e20210236. [Google Scholar] [CrossRef]
- Holbrook, M.R.; Shope, R.E.; Barrett, A.D.T. Use of recombinant E protein domain III-based enzyme-linked immunosorbent assays for differentiation of tick-borne encephalitis serocomplex flaviviruses from mosquito-borne flaviviruses. J. Clin. Microbiol. 2004, 42, 4101–4110. [Google Scholar] [CrossRef]
- Beasley, D.W.C.; Holbrook, M.R.; da Rosa, A.P.A.T.; Coffey, L.; Carrara, A.-S.; Phillippi-Falkenstein, K.; Bohm, R.P.; Ratterree, M.S.; Lillibridge, K.M.; Ludwig, G.V.; et al. Use of a recombinant envelope protein subunit antigen for specific serological diagnosis of west Nile Virus infection. J. Clin. Microbiol. 2004, 42, 2759–2765. [Google Scholar] [CrossRef]
- Reusken, C.; Boonstra, M.; Rugebregt, S.; Scherbeijn, S.; Chandler, F.; Avšič-Županc, T.; Vapalahti, O.; Koopmans, M.; GeurtsvanKessel, C.H. An evaluation of serological methods to diagnose tick-borne encephalitis from serum and cerebrospinal fluid. J. Clin. Virol. 2019, 120, 78–83. [Google Scholar] [CrossRef]
- Niedrig, M.; Vaisviliene, D.; Teichmann, A.; Klockmann, U.; Biel, S. Comparison of six different commercial IgG-ELISA kits for the detection of TBEV-antibodies. J. Clin. Virol. Off. Publ. Pan Am. Soc. Clin. Virol. 2001, 20, 179–182. [Google Scholar] [CrossRef]
- Papa, A.; Karabaxoglou, D.; Kansouzidou, A. Acute West Nile virus neuroinvasive infections: Cross-reactivity with dengue virus and tick-borne encephalitis virus. J. Med. Virol. 2011, 83, 1861–1865. [Google Scholar] [CrossRef] [PubMed]
- Cleton, N.; Godeke, G.-J.-J.; Reimerink, J.; Beersma, M.; van Doorn, H.R.; Franco, L.; Goeijenbier, M.; Jimenez-Clavero, M.A.; Johnson, B.W.; Niedrig, M.; et al. Spot the difference—Development of a syndrome based protein microarray for specific serological detection of multiple flavivirus infections in travelers. PLoS Neglected Trop. Dis. 2015, 9, e0003580. [Google Scholar] [CrossRef] [PubMed]
- Cleton, N.B.; van Maanen, K.; Bergervoet, S.A.; Bon, N.; Beck, C.; Godeke, G.-J.; Lecollinet, S.; Bowen, R.; Lelli, D.; Nowotny, N.; et al. A Serological Protein Microarray for Detection of Multiple Cross-Reactive Flavivirus Infections in Horses for Veterinary and Public Health Surveillance. Transbound. Emerg. Dis. 2017, 64, 1801–1812. [Google Scholar] [CrossRef] [PubMed]
- Thao, T.T.N.; de Bruin, E.; Phuong, H.T.; Vy, N.H.T.; Ham, H.-J.v.D.; Wills, B.A.; Tien, N.T.H.; Le Duyen, H.T.; Trung, D.T.; Whitehead, S.S.; et al. Using NS1 Flavivirus Protein Microarray to Infer Past Infecting Dengue Virus Serotype and Number of Past Dengue Virus Infections in Vietnamese Individuals. J. Infect. Dis. 2021, 223, 2053–2061. [Google Scholar] [CrossRef] [PubMed]
- Ndiaye, O.; Diagne, C.T.; El Wahed, A.A.; Dia, F.; Dia, M.; Faye, A.; Leal, S.D.V.; dos Santos, M.; Mendonça, M.d.L.d.L.; Leite, C.C.d.S.; et al. Use of Envelope Domain III Protein for the Detection of IgG Type Antibodies Specific to Zika Virus by Indirect ELISA. Diagnostics 2023, 13, 462. [Google Scholar] [CrossRef]
- Denis, J.; Attoumani, S.; Gravier, P.; Tenebray, B.; Garnier, A.; Briolant, S.; de Laval, F.; Chastres, V.; Grard, G.; Leparc-Goffart, I.; et al. High specificity and sensitivity of Zika EDIII-based ELISA diagnosis highlighted by a large human reference panel. PLoS Neglected Trop. Dis. 2019, 13, e0007747. [Google Scholar] [CrossRef] [PubMed]
- Premkumar, L.; Collins, M.; Graham, S.; Liou, G.-J.A.; Lopez, C.A.; Jadi, R.; Balmaseda, A.; Brackbill, J.A.; Dietze, R.; Camacho, E.; et al. Development of Envelope Protein Antigens to Serologically Differentiate Zika Virus Infection from Dengue Virus Infection. J. Clin. Microbiol. 2018, 56, e01504-17. [Google Scholar] [CrossRef] [PubMed]
- Binley, J.M.; Sanders, R.W.; Clas, B.; Schuelke, N.; Master, A.; Guo, Y.; Kajumo, F.; Anselma, D.J.; Maddon, P.J.; Olson, W.C.; et al. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J. Virol. 2000, 74, 627–643. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.-Y.; Song, W.-T.; Li, Y.; Chen, W.-J.; Yang, D.; Zhong, G.-C.; Zhou, H.-Z.; Ren, C.-Y.; Yu, H.-T.; Ling, H. Improved expression of secretory and trimeric proteins in mammalian cells via the introduction of a new trimer motif and a mutant of the tPA signal sequence. Appl. Microbiol. Biotechnol. 2011, 91, 731–740. [Google Scholar] [CrossRef] [PubMed]
- European Commission. Commission Implementing Decision (EU) 2018/945 of 22 June 2018 on the communicable diseases and related special health issues to be covered by epidemiological surveillance as well as relevant case definitions. OJEU 2018, 61, 170. [Google Scholar]
- Hofhuis, A.; Berg, O.v.D.; Meerstadt-Rombach, F.; Wijngaard, C.v.D.; Chung, N.; Franz, E.; Reimerink, J. Exposure to tick-borne encephalitis virus among nature management workers in the Netherlands. Ticks Tick-Borne Dis. 2021, 12, 101762. [Google Scholar] [CrossRef] [PubMed]
- Koopmans, M.; de Bruin, E.; Godeke, G.-J.; Friesema, I.; van Gageldonk, R.; Schipper, M.; Meijer, A.; van Binnendijk, R.; Rimmelzwaan, G.; de Jong, M.D.; et al. Profiling of humoral immune responses to influenza viruses by using protein microarray. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2012, 18, 797–807. [Google Scholar] [CrossRef] [PubMed]
- Reusken, C.; Mou, H.; Godeke, G.J.; van der Hoek, L.; Meyer, B.; A Müller, M.; Haagmans, B.; de Sousa, R.; Schuurman, N.; Dittmer, U.; et al. Specific serology for emerging human coronaviruses by protein microarray. Euro Surveill. Bull. Eur. Sur Mal. Transm. Eur. Commun. Dis. Bull. 2013, 18, 20441. [Google Scholar] [CrossRef] [PubMed]
- van Tol, S.; Mögling, R.; Li, W.; Godeke, G.-J.; Swart, A.; Bergmans, B.; Brandenburg, A.; Kremer, K.; Murk, J.-L.; van Beek, J.; et al. Accurate serology for SARS-CoV-2 and common human coronaviruses using a multiplex approach. Emerg. Microbes Infect. 2020, 9, 1965–1973. [Google Scholar] [CrossRef] [PubMed]
- Berrar, D.; Flach, P. Caveats and pitfalls of ROC analysis in clinical microarray research (and how to avoid them). Brief. Bioinform. 2012, 13, 83–97. [Google Scholar] [CrossRef]
- Pustijanac, E.; Buršić, M.; Talapko, J.; Škrlec, I.; Meštrović, T.; Lišnjić, D. Tick-Borne Encephalitis Virus: A Comprehensive Review of Transmission, Pathogenesis, Epidemiology, Clinical Manifestations, Diagnosis, and Prevention. Microorganisms 2023, 11, 1634. [Google Scholar] [CrossRef]
- da Silva, P.G.; dos Reis, J.A.S.; Rodrigues, M.N.; Ardaya, Q.d.S.; Mesquita, J.R. Serological Cross-Reactivity in Zoonotic Flaviviral Infections of Medical Importance. Antibodies 2023, 12, 18. [Google Scholar] [CrossRef]
- Makino, Y.; Tadano, M.; Saito, M.; Maneekarn, N.; Sittisombut, N.; Sirisanthana, V.; Poneprasert, B.; Fukunaga, T. Studies on serological cross-reaction in sequential flavivirus infections. Microbiol. Immunol. 1994, 38, 951–955. [Google Scholar] [CrossRef]
- Sánchez, M.D.; Pierson, T.C.; DeGrace, M.M.; Mattei, L.M.; Hanna, S.L.; Del Piero, F.; Doms, R.W. The neutralizing antibody response against West Nile virus in naturally infected horses. Virology 2007, 359, 336–348. [Google Scholar] [CrossRef]
- Svoboda, P.; Haviernik, J.; Bednar, P.; Matkovic, M.; Rincón, T.C.; Keeffe, J.; Palus, M.; Salat, J.; Agudelo, M.; Nussenzweig, M.C.; et al. A combination of two resistance mechanisms is critical for tick-borne encephalitis virus escape from a broadly neutralizing human antibody. Cell Rep. 2023, 42, 113149. [Google Scholar] [CrossRef]
- Matveeva, V.A.; Popova, R.V.; Kvetkova, E.A.; Chernicina, L.O.; Zlobin, V.I.; Puchovskaya, N.M.; Morozova, O.V. Antibodies against tick-borne encephalitis virus (TBEV) non-structural and structural proteins in human sera and spinal fluid. Immunol. Lett. 1995, 46, 1–4. [Google Scholar] [CrossRef]
- Erlanger, T.E.; Weiss, S.; Keiser, J.; Utzinger, J.; Wiedenmayer, K. Past, present, and future of japanese encephalitis. Emerg. Infect. Dis. 2009, 15, 1–7. [Google Scholar] [CrossRef]
- Platonov, A.E.; Rossi, G.; Karan, L.S.; O Mironov, K.; Busani, L.; Rezza, G. Does the Japanese encephalitis virus (JEV) represent a threat for human health in Europe? Detection of JEV RNA sequences in birds collected in Italy. Euro Surveill. Bull. Eur. Sur Mal. Transm. Eur. Commun. Dis. Bull. 2012, 17, 20241. [Google Scholar] [CrossRef] [PubMed]
- Zaaijer, H.L.; Slot, E.; Molier, M.; Reusken, C.B.; Koppelman, M.H. Usutu virus infection in Dutch blood donors. Transfusion 2019, 59, 2931–2937. [Google Scholar] [CrossRef] [PubMed]
- Rijks, J.M.; Kik, M.L.; Slaterus, R.; Foppen, R.; Stroo, A.; Ijzer, J.; Stahl, J.; Gröne, A.; Koopmans, M.; van der Jeugd, H.P.; et al. Widespread Usutu virus outbreak in birds in the Netherlands, 2016. Eurosurveillance 2016, 21, 30391. [Google Scholar] [CrossRef] [PubMed]
- Grobusch, M.P.; Weld, L.; Goorhuis, A.; Hamer, D.H.; Schunk, M.; Jordan, S.; Mockenhaupt, F.P.; Chappuis, F.; Asgeirsson, H.; Caumes, E.; et al. Travel-related infections presenting in Europe: A 20-year analysis of EuroTravNet surveillance data. Lancet Reg. Health Eur. 2021, 1, 100001. [Google Scholar] [CrossRef] [PubMed]
- Prates, J.W.O.; Xisto, M.F.; Rodrigues, J.V.d.S.; Colombari, J.P.C.; Meira, J.M.A.; Dias, R.S.; da Silva, C.C.; de Paula, E.S.O. Zika Virus Envelope Protein Domain III Produced in K. phaffii Has the Potential for Diagnostic Applications. Diagnostics 2022, 12, 1198. [Google Scholar] [CrossRef] [PubMed]
- Campos, G.S.; Bandeira, A.C.; Sardi, S.I. Zika Virus Outbreak, Bahia, Brazil. Emerg. Infect. Dis. 2015, 21, 1885–1886. [Google Scholar] [CrossRef]
- Kuno, G. Serodiagnosis of flaviviral infections and vaccinations in humans. Adv. Virus Res. 2003, 61, 3–65. [Google Scholar] [CrossRef] [PubMed]
- Gromowski, G.D.; Barrett, A.D. Characterization of an antigenic site that contains a dominant, type-specific neutralization determinant on the envelope protein domain III (ED3) of dengue 2 virus. Virology 2007, 366, 349–360. [Google Scholar] [CrossRef]
- Sankar, S.G.; Balaji, T.; Venkatasubramani, K.; Thenmozhi, V.; Dhananjeyan, K.; Paramasivan, R.; Tyagi, B.; Vennison, S.J. Dengue NS1 and prM antibodies increase the sensitivity of acute dengue diagnosis test and differentiate from Japanese encephalitis infection. J. Immunol. Methods 2014, 407, 116–119. [Google Scholar] [CrossRef]
- Hall, R.A.; Kay, B.H.; Burgess, G.W.; Clancy, P.; Fanning, I.D. Epitope analysis of the envelope and non-structural glycoproteins of Murray Valley encephalitis virus. J. Gen. Virol. 1990, 71 Pt 12, 2923–2930. [Google Scholar] [CrossRef]
- Falconar, A.K.; Young, P.R. Immunoaffinity purification of native dimer forms of the flavivirus non-structural glycoprotein, NS1. J. Virol. Methods 1990, 30, 323–332. [Google Scholar] [CrossRef] [PubMed]
- Albinsson, B.; Rönnberg, B.; Vene, S.; Lundkvist, Å. Antibody responses to tick-borne encephalitis virus non-structural protein 1 and whole virus antigen—A new tool in the assessment of suspected vaccine failure patients. Infect. Ecol. Epidemiology 2019, 9, 1696132. [Google Scholar] [CrossRef] [PubMed]
- Stiasny, K.; Leitner, A.; Holzmann, H.; Heinz, F.X. Dynamics and Extent of Non-Structural Protein 1-Antibody Responses in Tick-Borne Encephalitis Vaccination Breakthroughs and Unvaccinated Patients. Viruses 2021, 13, 1007. [Google Scholar] [CrossRef]
- Albinsson, B.; Vene, S.; Rombo, L.; Blomberg, J.; Lundkvist, Å.; Rönnberg, B. Distinction between serological responses following tick-borne encephalitis virus (TBEV) infection vs vaccination, Sweden 2017. Euro Surveill. Bull. Eur. Sur Mal. Transm. Eur. Commun. Dis. Bull. 2018, 23, 17–00838. [Google Scholar] [CrossRef]
- Girl, P.; Bestehorn-Willmann, M.; Zange, S.; Borde, J.P.; Dobler, G.; von Buttlar, H. Tick-Borne Encephalitis Virus Nonstructural Protein 1 IgG Enzyme-Linked Immunosorbent Assay for Differentiating Infection versus Vaccination Antibody Responses. J. Clin. Microbiol. 2020, 58, e01783-19. [Google Scholar] [CrossRef]
- Dobler, G.; Euringer, K.; Kaier, K.; Borde, J.P. Serological Protection Rates against TBEV Infection in Blood Donors from a Highly Endemic Region in Southern Germany. Vaccines 2023, 11, 522. [Google Scholar] [CrossRef] [PubMed]
- Salat, J.; Mikulasek, K.; Larralde, O.; Formanova, P.P.; Chrdle, A.; Haviernik, J.; Elsterova, J.; Teislerova, D.; Palus, M.; Eyer, L.; et al. Tick-Borne Encephalitis Virus Vaccines Contain Non-Structural Protein 1 Antigen and May Elicit NS1-Specific Antibody Responses in Vaccinated Individuals. Vaccines 2020, 8, 81. [Google Scholar] [CrossRef] [PubMed]


| Virus Species | Number of Samples | Sex | Group Age (in Years) | Positive Serology (ELISA/IFA) Specific IgG | Virus Neutralization Confirmed (VNT/PRTN) | Vaccination |
|---|---|---|---|---|---|---|
| DENV1–4 | 11 | M (6/11) F (5/11) | 19 to 70 | 11/11 | N/D | N/A |
| ZIKV | 15 | M (3/15) F (12/15) | 26 to 80 | 15/15 (CHIKV and DENV positive IgG IFA: 7/15) | N/D | N/A |
| WNV | 7 | M (3/7) F (4/7) | 30 to 75 | 7/7 | 5/7 | N/A |
| TBEV | 20 | M (9/20) F (8/20) | 20 to 80 | 20/20 | 15/20 | 9/20 N/A 10/20 No vaccine 1/20 YF vaccine |
| TBEV Vaccinated group | 13 | M (4/13) F (9/13) | 17 to 70 | 13/13 | N/D | TBEV vaccine * |
| Negative group | 35 | N/A | N/A | Orthoflavivirus IgM/IgG Neg | N/D | N/A |
| Dutch forestry workers | 556 | N/A | 22 to 88 | TBEV IgG (10/556) | TBEV VNT (3/10) | No vaccine |
| Microarray Value | |||||
|---|---|---|---|---|---|
| Sample | NS1 TBEV | EDIII TBEV | TBEV ELISA Value * | VNT Titers * | Interpretation |
| 1 | ≤10 | 218 | 6.1 | 120 | Confirmed TBEV infection |
| 2 | ≤10 | 126 | 3.0 | 30 | Confirmed TBEV infection |
| 3 | ≤10 | 101 | 1.3 | 30 | Confirmed TBEV infection |
| 4 | ≤10 | 36 | 2.6 | Neg | Possible TBEV infection |
| 5 | ≤10 | 40 | 2.4 | Neg | Possible TBEV infection |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Valle, C.; Shrestha, S.; Godeke, G.-J.; Hoogerwerf, M.N.; Reimerink, J.; Eggink, D.; Reusken, C. Multiplex Serology for Sensitive and Specific Flavivirus IgG Detection: Addition of Envelope Protein Domain III to NS1 Increases Sensitivity for Tick-Borne Encephalitis Virus IgG Detection. Viruses 2024, 16, 286. https://doi.org/10.3390/v16020286
Valle C, Shrestha S, Godeke G-J, Hoogerwerf MN, Reimerink J, Eggink D, Reusken C. Multiplex Serology for Sensitive and Specific Flavivirus IgG Detection: Addition of Envelope Protein Domain III to NS1 Increases Sensitivity for Tick-Borne Encephalitis Virus IgG Detection. Viruses. 2024; 16(2):286. https://doi.org/10.3390/v16020286
Chicago/Turabian StyleValle, Coralie, Sandhya Shrestha, Gert-Jan Godeke, Marieke N. Hoogerwerf, Johan Reimerink, Dirk Eggink, and Chantal Reusken. 2024. "Multiplex Serology for Sensitive and Specific Flavivirus IgG Detection: Addition of Envelope Protein Domain III to NS1 Increases Sensitivity for Tick-Borne Encephalitis Virus IgG Detection" Viruses 16, no. 2: 286. https://doi.org/10.3390/v16020286





