Identification and Genomic Characterization of Bovine Boosepivirus A in the United States and Canada
Abstract
:1. Introduction
2. Materials and Methods
2.1. Samples
2.2. RNA Extraction
2.3. Laboratory Testing
2.4. Next-Generation Sequencing (NGS)
2.5. Bioinformatic Analysis
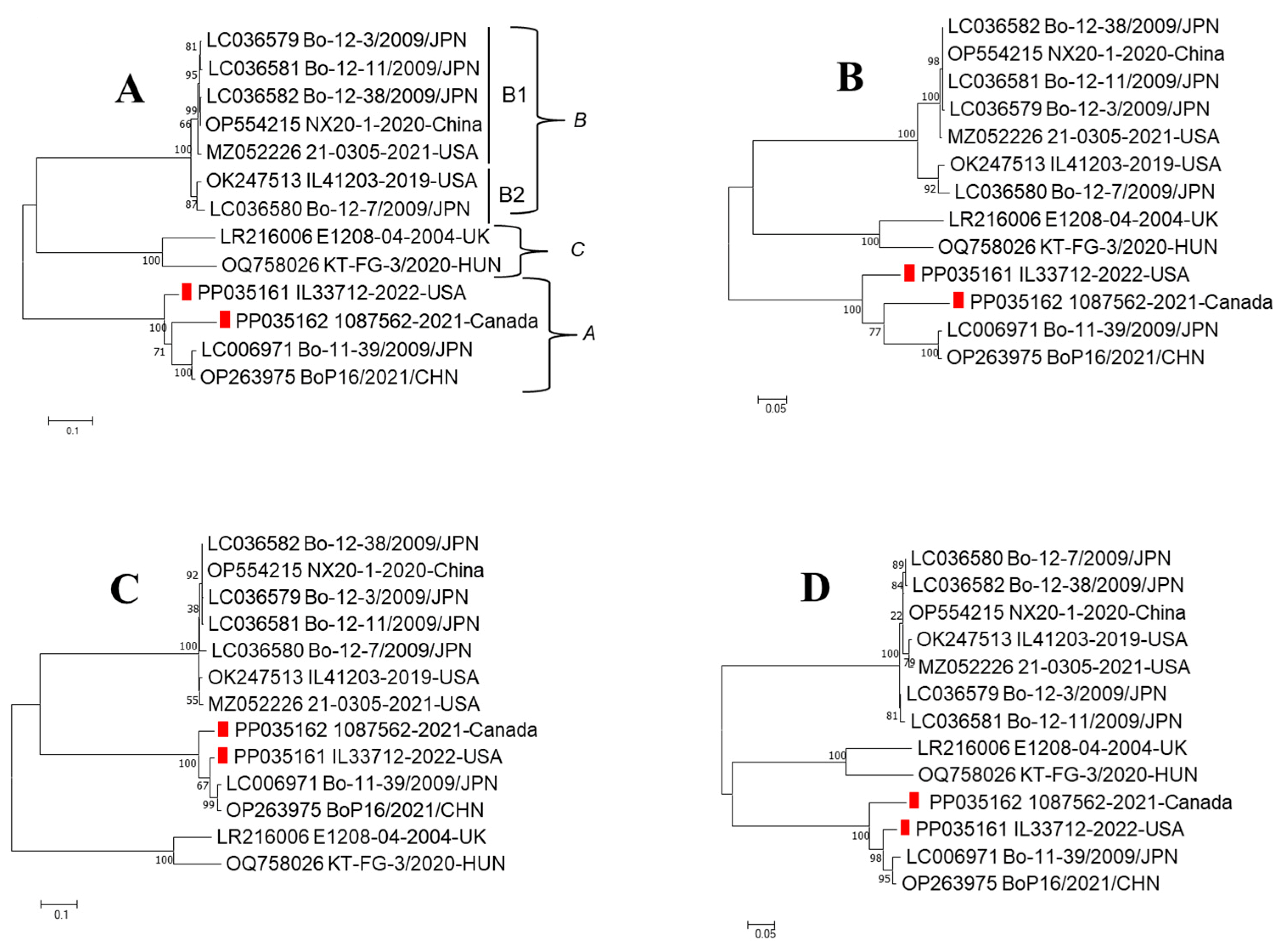
3. Results
3.1. Routine Laboratory Testing Result
3.2. NGS
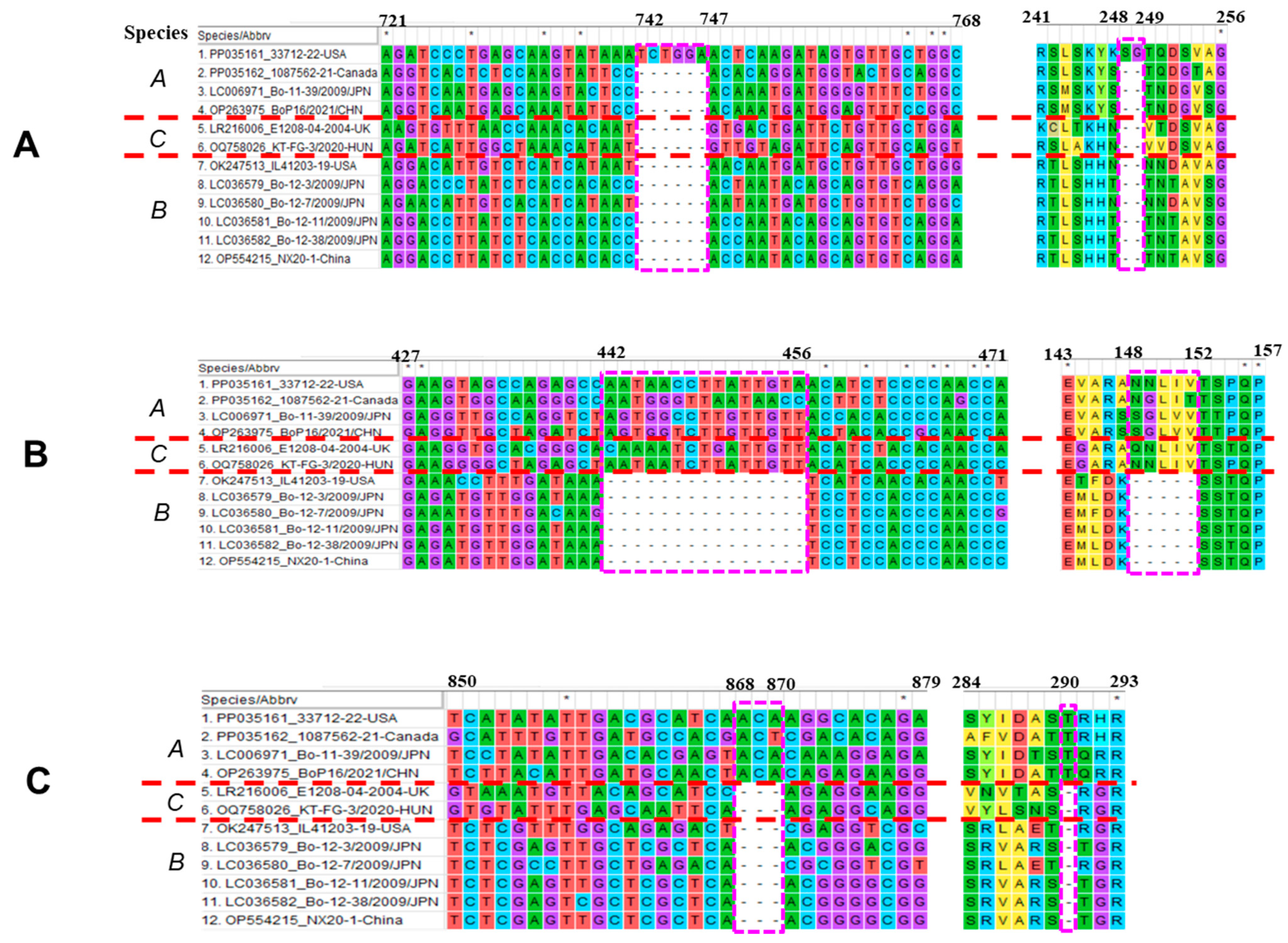
3.3. Genome Characterization of Boosepivirus
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Correction Statement
References
- Zell, R.; Delwart, E.; Gorbalenya, A.E.; Hovi, T.; King, A.M.Q.; Knowles, N.J.; Lindberg, A.M.; Pallansch, M.A.; Palmenberg, A.C.; Reuter, G.; et al. ICTV Virus Taxonomy Profile: Picornaviridae. J. Gen. Virol. 2017, 98, 2421–2422. [Google Scholar] [CrossRef] [PubMed]
- Xue, M.; Wang, H.; Li, W.; Zhou, G.; Tu, Y.; Yu, L. Effects of amino acid substitutions in the VP2 B-C loop on antigenicity and pathogenicity of serotype Asia1 foot-and-mouth disease virus. Virol. J. 2012, 9, 191. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Yan, M.; Hao, J.; Shen, C.; Zhu, Z.; Zhang, D.; Hou, J.; Xu, G.; Li, D.; Zheng, H.; et al. Foot-and-Mouth Disease Virus Structural Protein VP1 Destroys the Stability of TPL2 Trimer by Degradation TPL2 to Evade Host Antiviral Immunity. J. Virol. 2021, 95, 7. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Lim, A.; Fredrickson, R. Genomic characterization of a new bovine picornavirus (boosepivirus) in diarrheal cattle and detection in different states of the United States, 2019. Transbound. Emerg. Dis. 2022, 69, 3109–3114. [Google Scholar] [CrossRef] [PubMed]
- Zell, R.; Knowles, N.J.; Simmonds, P. A proposed division of the family Picornaviridae into subfamilies based on phylogenetic relationships and functional genomic organization. Arch. Virol. 2021, 166, 2927–2935. [Google Scholar] [CrossRef] [PubMed]
- ICTV Family: Picornaviridae, Genus: Boosepivirus. Available online: https://ictv.global/report/chapter/picornaviridae/picornaviridae/boosepivirus (accessed on 28 December 2023).
- Nagai, M.; Omatsu, T.; Aoki, H.; Kaku, Y.; Belsham, G.J.; Haga, K.; Naoi, Y.; Sano, K.; Umetsu, M.; Shiokawa, M.; et al. Identification and complete genome analysis of a novel bovine picornavirus in Japan. Virus Res. 2015, 210, 205–212. [Google Scholar] [CrossRef] [PubMed]
- Ji, C.; Zhang, Y.; Feng, Y.; Zhang, X.; Ma, J.; Pan, Z.; Kawaguchi, A.; Yao, H. Systematic Surveillance of an Emerging Picornavirus among Cattle and Sheep in China. Microbiol. Spectr. 2023, 11, e0504022. [Google Scholar] [CrossRef] [PubMed]
- Forth, L.F.; Scholes, S.F.E.; Pesavento, P.A.; Jackson, K.; Mackintosh, A.; Carson, A.; Howie, F.; Schlottau, K.; Wernike, K.; Pohlmann, A.; et al. Novel Picornavirus in Lambs with Severe Encephalomyelitis. Emerg. Infect. Dis. 2019, 25, 963–967. [Google Scholar] [CrossRef] [PubMed]
- Boros, A.; Pankovics, P.; Laszlo, Z.; Urban, P.; Herczeg, R.; Gaspar, G.; Toth, F.; Reuter, G. The genomic and epidemiological investigations of enteric viruses of domestic caprine (Capra hircus) revealed the presence of multiple novel viruses related to known strains of humans and ruminant livestock species. Microbiol. Spectr. 2023, 11, e0253323. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Stuber, T.; Camp, P.; Robbe-Austerman, S.; Zhang, Y. Whole-Genome Sequencing of Porcine Epidemic Diarrhea Virus by Illumina MiSeq Platform. In Animal Coronaviruses; Wang, L., Ed.; Springer: Berlin/Heidelberg, Germany, 2016; pp. 201–208. [Google Scholar]
- Prjibelski, A.; Antipov, D.; Meleshko, D.; Lapidus, A.; Korobeynikov, A. Using SPAdes De Novo Assembler. Curr. Protoc. Bioinform. 2020, 70, e102. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Zheng, W.; Li, Y.; Pearce, R.; Zhang, C.; Bell, E.W.; Zhang, G.; Zhang, Y. I-TASSER-MTD: A deep-learning-based platform for multi-domain protein structure and function prediction. Nat. Protoc. 2022, 17, 2326–2353. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Fredrickson, R.; Duncan, M.; Samuelson, J.; Hsiao, S.H. Bovine Kobuvirus in Calves with Diarrhea, United States. Emerg. Infect. Dis. 2020, 26, 176–178. [Google Scholar] [CrossRef] [PubMed]
- Hao, L.; Chen, C.; Bailey, K.; Wang, L. Bovine kobuvirus-A comprehensive review. Transbound. Emerg. Dis. 2021, 68, 1886–1894. [Google Scholar] [CrossRef] [PubMed]
- Salles, M.W.; Scholes, S.F.; Dauber, M.; Strebelow, G.; Wojnarowicz, C.; Hassard, L.; Acton, A.C.; Bollinger, T.K. Porcine teschovirus polioencephalomyelitis in western Canada. J. Vet. Diagn. Invest. 2011, 23, 367–373. [Google Scholar] [CrossRef] [PubMed]
- Rossi, T.M.; Moore, A.; O’Sullivan, T.L.; Greer, A.L. Risk factors for duration of equine rhinitis A virus respiratory disease. Equine Vet. J. 2020, 52, 369–373. [Google Scholar] [CrossRef] [PubMed]
- Brewer, L.A.; Lwamba, H.C.; Murtaugh, M.P.; Palmenberg, A.C.; Brown, C.; Njenga, M.K. Porcine encephalomyocarditis virus persists in pig myocardium and infects human myocardial cells. J. Virol. 2001, 75, 11621–11629. [Google Scholar] [CrossRef] [PubMed]


| A | ||||||||||||||||
| PP035161_IL33712-2022-USA | Complete * | Polyprotein | P1 | P2 | P3 | VP4 | VP2 | VP3 | VP1 | 2A | 2B | 2C | 3A | 3B | 3C | 3D |
| OP263975_BoP16/2021/CHN | 77.6 | 90.4 | 81.7 | 95.5 | 95.5 | 98.4 | 80.0 | 84.4 | 77.6 | 90.3 | 99.0 | 97.5 | 96.0 | 100 | 95.5 | 95.2 |
| LC006971_Bo-11-39/2009/JPN | 77.5 | 90.2 | 81.6 | 95.8 | 94.7 | 96.9 | 80.4 | 84.0 | 77.6 | 91.3 | 99.0 | 97.5 | 96.0 | 86.9 | 96.1 | 94.3 |
| PP035162_1087562-2021-Canada | 75.0 | 86.1 | 80.5 | 91.6 | 88.5 | 84.6 | 81.6 | 86.6 | 74.4 | 85.9 | 93.3 | 94.5 | 82.0 | 100 | 85.0 | 90.7 |
| LR216006_E1208-04-2004-UK | 54.8 | 52.4 | 56.8 | 42.1 | 59.8 | 64.6 | 54.7 | 60.3 | 55.3 | 29 | 38.8 | 51.1 | 46.0 | 52.1 | 64.8 | 61.0 |
| OQ758026_KT-FG-3/2020-HUN | 56.6 | 51.4 | 55.7 | 41.8 | 58.8 | 64.6 | 56.8 | 57.2 | 52.8 | 25.7 | 43.5 | 51.4 | 44.0 | 47.8 | 58.7 | 61.6 |
| OK247513_IL41203-2019-USA | 55.1 | 52.2 | 55.2 | 47.7 | 56.2 | 61.7 | 57.9 | 58.1 | 50.4 | 33.9 | 34.8 | 59.9 | 38.0 | 43.4 | 54.9 | 63.0 |
| LC036579_Bo-12-3/2009/JPN | 54.5 | 52.2 | 55.1 | 47.7 | 56.4 | 61.7 | 57.9 | 57.7 | 49.2 | 33.9 | 34.8 | 59.6 | 38.0 | 43.4 | 54.9 | 63.2 |
| LC036580_Bo-12-7/2009/JPN | 54.7 | 52.1 | 55.6 | 46.8 | 56.2 | 61.7 | 57.9 | 59.0 | 50.4 | 33.0 | 34.8 | 59.3 | 38.0 | 43.4 | 54.9 | 63.0 |
| MZ052226_21-0305-USA | - | 52.0 | 55.0 | 47.7 | 56.2 | 61.7 | 58.3 | 57.2 | 49.2 | 33.4 | 34.8 | 59.9 | 38.0 | 43.4 | 55.4 | 62.7 |
| LC036581_Bo-12-11/2009/JPN | 54.0 | 52.1 | 55.1 | 47.9 | 56.2 | 61.7 | 57.9 | 57.7 | 49.2 | 33.9 | 34.8 | 59.6 | 38.0 | 43.4 | 54.9 | 63.0 |
| LC036582_Bo-12-38/2009/JPN | 54.3 | 52.1 | 55.1 | 47.9 | 56.4 | 61.7 | 57.9 | 57.7 | 49.2 | 33.9 | 34.8 | 59.6 | 38.0 | 43.4 | 54.9 | 63.2 |
| OP554215_NX20-1-2020-China | 54.4 | 52.2 | 55.1 | 47.9 | 56.4 | 61.7 | 57.9 | 57.7 | 49.2 | 33.9 | 34.8 | 59.6 | 38.0 | 43.4 | 54.9 | 63.2 |
| B | ||||||||||||||||
| PP035162_1087562-2021-Canada | Complete | Polyprotein | P1 | P2 | P3 | VP4 | VP2 | VP3 | VP1 | 2A | 2B | 2C | 3A | 3B | 3C | 3D |
| PP035161_IL33712-2022-USA | 75.0 | 86.1 | 80.5 | 91.6 | 88.5 | 84.6 | 81.6 | 86.6 | 74.4 | 85.9 | 93.3 | 94.5 | 82.0 | 100 | 85.0 | 90.7 |
| OP263975_BoP16/2021/CHN | 74.1 | 86.1 | 80.9 | 90.2 | 89.0 | 84.6 | 81.8 | 85.7 | 75.8 | 84.0 | 92.4 | 93.3 | 82.0 | 100 | 84.5 | 91.8 |
| LC006971_Bo-11-39/2009/JPN | 74.0 | 85.8 | 80.8 | 89.7 | 88.8 | 86.1 | 81.8 | 85.3 | 75.5 | 82.6 | 92.4 | 93.3 | 84.0 | 86.9 | 85.6 | 91.1 |
| LR216006_E1208-04-2004-UK | 53.6 | 51.2 | 54.4 | 41.7 | 59.5 | 60.0 | 52.9 | 62.1 | 50.1 | 28.5 | 38.8 | 50.8 | 45.0 | 52.1 | 65.3 | 60.3 |
| OQ758026_KT-FG-3/2020-HUN | 55.1 | 50.7 | 54.2 | 42.1 | 57.9 | 61.5 | 55.0 | 60.3 | 49.3 | 25.2 | 42.5 | 53.2 | 41.0 | 47.8 | 59.3 | 60.5 |
| OK247513_IL41203-2019-USA | 53.2 | 51.2 | 53.0 | 47.4 | 56.0 | 57.3 | 55.2 | 59.4 | 46.6 | 32.0 | 33.9 | 60.5 | 38.0 | 43.4 | 58.7 | 61.0 |
| LC036579_Bo-12-3/2009/JPN | 53.6 | 51.4 | 53.4 | 47.7 | 56.1 | 57.3 | 55.6 | 58.5 | 47.5 | 33.0 | 33.9 | 60.2 | 37.3 | 43.4 | 58.7 | 61.5 |
| LC036580_Bo-12-7/2009/JPN | 53.4 | 51.3 | 53.1 | 47.1 | 56 | 57.3 | 54.8 | 59.0 | 47.2 | 33.0 | 33.9 | 60.5 | 37.3 | 43.4 | 58.7 | 61.2 |
| MZ052226_21-0305-USA | - | 51.1 | 53.1 | 47.4 | 55.9 | 57.3 | 56.1 | 58.1 | 46.6 | 31.6 | 33.9 | 60.5 | 37.3 | 43.4 | 59.3 | 60.8 |
| LC036581_Bo-12-11/2009/JPN | 53.0 | 51.3 | 53.3 | 47.6 | 56.0 | 57.3 | 55.6 | 58.5 | 47.2 | 33.0 | 33.9 | 60.2 | 37.3 | 43.4 | 58.2 | 61.5 |
| LC036582_Bo-12-38/2009/JPN | 53.3 | 51.3 | 53.3 | 47.6 | 56.0 | 57.3 | 55.6 | 58.5 | 47.2 | 33.0 | 33.9 | 60.2 | 37.3 | 43.4 | 58.7 | 61.2 |
| OP554215_NX20-1-2020-China | 53.5 | 51.3 | 53.3 | 47.6 | 56.1 | 56.7 | 55.6 | 58.5 | 47.2 | 33.0 | 33.9 | 60.2 | 37.3 | 43.4 | 58.7 | 61.5 |
| C | ||||||||||||||||
| OP263975_BoP16/2021/CHN | Complete | Polyprotein | P1 | P2 | P3 | VP4 | VP2 | VP3 | VP1 | 2A | 2B | 2C | 3A | 3B | 3C | 3D |
| LC006971_Bo-11-39/2009/JPN | 86.7 | 98.3 | 99.0 | 98.1 | 97.9 | 98.4 | 99.5 | 99.5 | 98.3 | 94.2 | 100 | 100 | 98.0 | 86.9 | 97.2 | 98.7 |
| PP035161_IL33712-2022-USA | 77.6 | 90.4 | 81.7 | 95.5 | 95.5 | 98.4 | 80.0 | 84.4 | 77.6 | 90.3 | 99.0 | 97.5 | 96.0 | 100 | 95.5 | 95.2 |
| PP035162_1087562-2021-Canada | 74.1 | 86.1 | 80.9 | 90.2 | 89.0 | 84.6 | 81.8 | 85.7 | 75.8 | 84.0 | 92.4 | 93.3 | 82.0 | 100 | 84.5 | 91.8 |
| LR216006_E1208-04-2004-UK | 56.0 | 52.1 | 55.5 | 41.8 | 60.7 | 64.6 | 54.6 | 62.9 | 49.8 | 28.5 | 37.9 | 51.4 | 46.0 | 52.1 | 65.3 | 62.3 |
| OQ758026_KT-FG-3/2020-HUN | 56.2 | 51.1 | 55.1 | 40.9 | 59.1 | 64.6 | 54.6 | 59.9 | 50.6 | 25.2 | 42.5 | 50.8 | 44.0 | 47.8 | 57.6 | 62.7 |
| OK247513_IL41203-2019-USA | 54.5 | 51.6 | 54.6 | 47.3 | 55.6 | 61.7 | 54.4 | 59.4 | 50.4 | 31.6 | 35.7 | 60.5 | 36.5 | 43.4 | 54.9 | 62.3 |
| LC036579_Bo-12-3/2009/JPN | 54.5 | 51.6 | 54.3 | 47.6 | 55.6 | 61.7 | 54.8 | 59.4 | 48.5 | 32.5 | 35.7 | 60.2 | 36.5 | 43.4 | 54.9 | 62.3 |
| LC036580_Bo-12-7/2009/JPN | 54.9 | 51.8 | 55.2 | 46.5 | 55.5 | 61.7 | 55.2 | 59.0 | 51.7 | 31.6 | 35.7 | 59.9 | 36.5 | 43.4 | 54.9 | 62.1 |
| MZ052226_21-0305-USA | - | 51.5 | 54.3 | 47.3 | 55.6 | 61.7 | 54.8 | 59.4 | 48.8 | 31.1 | 35.7 | 60.5 | 36.5 | 43.4 | 55.4 | 62.1 |
| LC036581_Bo-12-11/2009/JPN | 54.0 | 51.7 | 54.5 | 47.4 | 55.6 | 61.7 | 54.8 | 59.4 | 48.8 | 32.5 | 35.7 | 60.2 | 36.5 | 43.4 | 54.9 | 62.3 |
| LC036582_Bo-12-38/2009/JPN | 54.3 | 51.6 | 54.5 | 47.4 | 55.6 | 61.7 | 54.8 | 59.4 | 48.8 | 32.5 | 35.7 | 60.2 | 36.5 | 43.4 | 54.9 | 62.3 |
| OP554215_NX20-1-2020-China | 54.4 | 51.7 | 54.5 | 47.4 | 55.6 | 61.7 | 54.8 | 59.4 | 48.8 | 32.5 | 35.7 | 60.2 | 36.5 | 43.4 | 54.9 | 62.3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Savard, C.; Wang, L. Identification and Genomic Characterization of Bovine Boosepivirus A in the United States and Canada. Viruses 2024, 16, 307. https://doi.org/10.3390/v16020307
Savard C, Wang L. Identification and Genomic Characterization of Bovine Boosepivirus A in the United States and Canada. Viruses. 2024; 16(2):307. https://doi.org/10.3390/v16020307
Chicago/Turabian StyleSavard, Christian, and Leyi Wang. 2024. "Identification and Genomic Characterization of Bovine Boosepivirus A in the United States and Canada" Viruses 16, no. 2: 307. https://doi.org/10.3390/v16020307






