Hepatitis E Virus in Domestic Ruminants and Virus Excretion in Milk—A Potential Source of Zoonotic HEV Infection
Abstract
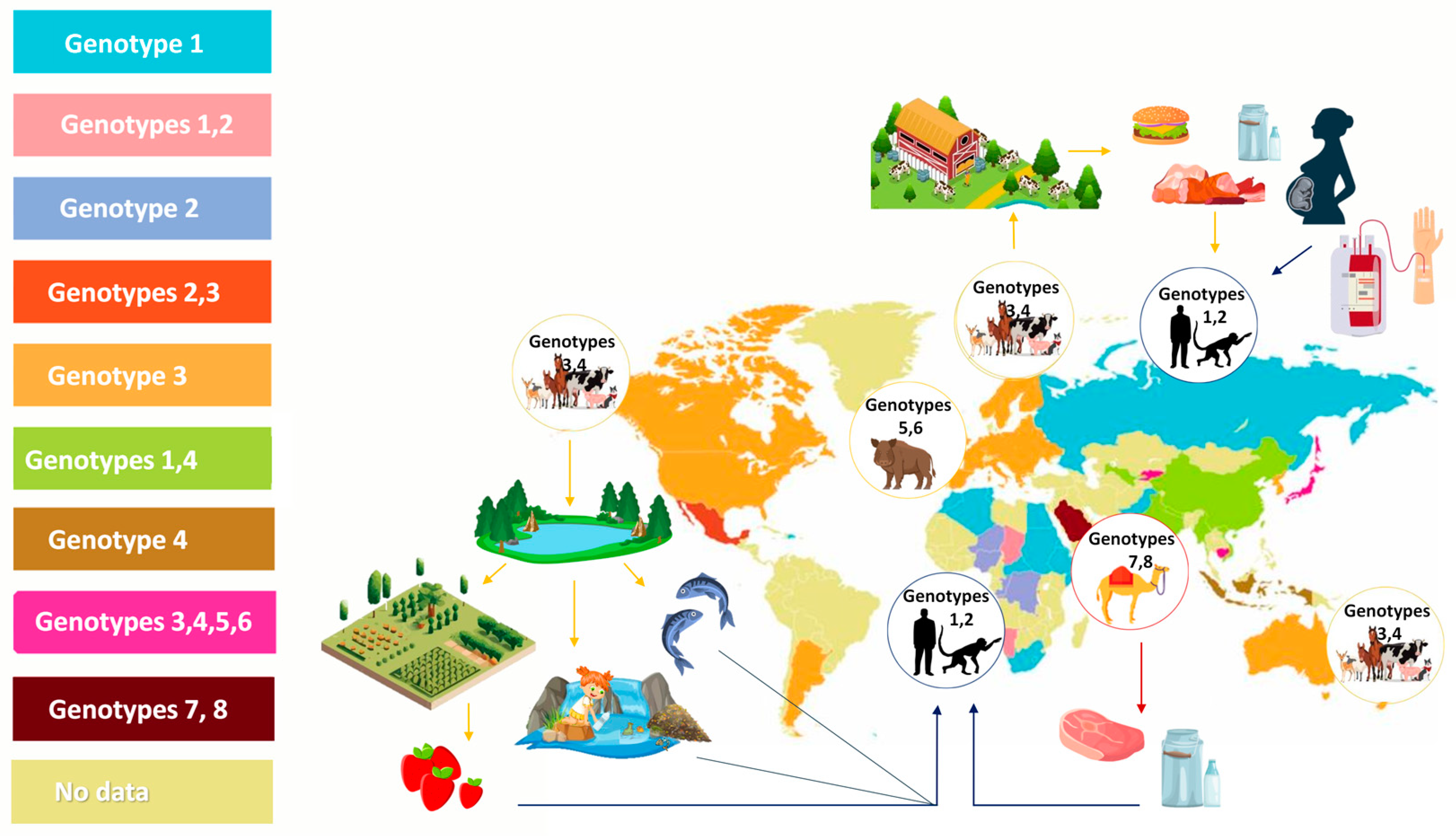
:1. Introduction
2. Hepatitis E Virus in Domestic Ruminants
2.1. Seroprevalence of Hepatitis E Virus in Ruminants
2.2. HEV RNA Prevalence in Domestic Ruminants
2.2.1. HEV in Goats
2.2.2. HEV in Sheep
2.2.3. HEV in Bovines, Genus Bos with the Spotlight on Cow Milk
3. Significance of HEV in Milk
4. Milk and Milk Safety Measures to Prevent Foodborne Zoonotic HEV Transmission
5. Hepatitis E Virus Cross-Species Transmission
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lozano, R.; Naghavi, M.; Foreman, K.; Lim, S.; Shibuya, K.; Aboyans, V.; Abraham, J.; Adair, T.; Aggarwal, R.; Ahn, S.Y.; et al. Global and Regional Mortality from 235 Causes of Death for 20 Age Groups in 1990 and 2010: A Systematic Analysis for the Global Burden of Disease Study 2010. Lancet 2012, 380, 2095–2128. [Google Scholar] [CrossRef] [PubMed]
- Rein, D.B.; Stevens, G.A.; Theaker, J.; Wittenborn, J.S.; Wiersma, S.T. The Global Burden of Hepatitis E Virus Genotypes 1 and 2 in 2005. Hepatology 2012, 55, 988–997. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Liu, J.; Li, Y.; Su, J.; Ma, Z.; Bramer, W.M.; Cao, W.; de Man, R.A.; Peppelenbosch, M.P.; Pan, Q. The Global Epidemiology of Hepatitis E Virus Infection: A Systematic Review and Meta-Analysis. Liver Int. 2020, 40, 1516–1528. [Google Scholar] [CrossRef] [PubMed]
- Takakusagi, S.; Kakizaki, S.; Takagi, H. The Diagnosis, Pathophysiology, and Treatment of Chronic Hepatitis E Virus Infection—A Condition Affecting Immunocompromised Patients. Microorganisms 2023, 11, 1303. [Google Scholar] [CrossRef]
- Damiris, K.; Meybodi, M.A.; Niazi, M.; Pyrsopoulos, N. Hepatitis E in Immunocompromised Individuals. World J. Hepatol. 2022, 14, 482–494. [Google Scholar] [CrossRef]
- Bergløv, A.; Hallager, S.; Weis, N. Hepatitis E during Pregnancy: Maternal and Foetal Case-Fatality Rates and Adverse Outcomes—A Systematic Review. J. Viral Hepat. 2019, 26, 1240–1248. [Google Scholar] [CrossRef]
- Khuroo, M.S. Discovery of Hepatitis E and Its Impact on Global Health: A Journey of 44 Years about an Incredible Human-Interest Story. Viruses 2023, 15, 1745. [Google Scholar] [CrossRef] [PubMed]
- Grange, Z.L.; Goldstein, T.; Johnson, C.K.; Anthony, S.; Gilardi, K.; Daszak, P.; Olival, K.J.; O’Rourke, T.; Murray, S.; Olson, S.H.; et al. Ranking the Risk of Animal-to-Human Spillover for Newly Discovered Viruses. Proc. Natl. Acad. Sci. USA 2021, 118, e2002324118. [Google Scholar] [CrossRef] [PubMed]
- European Food Safety Authority (EFSA); Berezowski, J.; de Balogh, K.; Dórea, F.C.; Rüegg, S.; Broglia, A.; Gervelmeyer, A.; Kohnle, L. Prioritisation of Zoonotic Diseases for Coordinated Surveillance Systems under the One Health Approach for Cross-Border Pathogens That Threaten the Union. EFSA J. 2023, 21, e07853. [Google Scholar] [CrossRef]
- Meng, X.J. Hepatitis E Virus: Animal Reservoirs and Zoonotic Risk. Vet. Microbiol. 2010, 140, 256–265. [Google Scholar] [CrossRef]
- Pavio, N.; Doceul, V.; Bagdassarian, E.; Johne, R. Recent Knowledge on Hepatitis E Virus in Suidae Reservoirs and Transmission Routes to Human. Vet. Res. 2017, 48, 78. [Google Scholar] [CrossRef] [PubMed]
- Hewitt, P.E.; Ijaz, S.; Brailsford, S.R.; Brett, R.; Dicks, S.; Haywood, B.; Kennedy, I.T.R.; Kitchen, A.; Patel, P.; Poh, J.; et al. Hepatitis E Virus in Blood Components: A Prevalence and Transmission Study in Southeast England. Lancet 2014, 384, 1766–1773. [Google Scholar] [CrossRef] [PubMed]
- Kuniholm, M.H.; Purcell, R.H.; McQuillan, G.M.; Engle, R.E.; Wasley, A.; Nelson, K.E. Epidemiology of Hepatitis E Virus in the United States: Results from the Third National Health and Nutrition Examination Survey, 1988–1994. J. Infect. Dis. 2009, 200, 48–56. [Google Scholar] [CrossRef] [PubMed]
- Mansuy, J.M.; Gallian, P.; Dimeglio, C.; Saune, K.; Arnaud, C.; Pelletier, B.; Morel, P.; Legrand, D.; Tiberghien, P.; Izopet, J. A Nationwide Survey of Hepatitis E Viral Infection in French Blood Donors. Hepatology 2016, 63, 1145–1154. [Google Scholar] [CrossRef]
- Purdy, M.A.; Drexler, J.F.; Meng, X.-J.; Norder, H.; Okamoto, H.; Van der Poel, W.H.M.; Reuter, G.; de Souza, W.M.; Ulrich, R.G.; Smith, D.B. ICTV Virus Taxonomy Profile: Hepeviridae 2022. J. Gen. Virol. 2022, 103, 001778. [Google Scholar] [CrossRef]
- Wang, B.; Meng, X.-J. Structural and Molecular Biology of Hepatitis E Virus. Comput. Struct. Biotechnol. J. 2021, 19, 1907–1916. [Google Scholar] [CrossRef]
- Nair, V.P.; Anang, S.; Subramani, C.; Madhvi, A.; Bakshi, K.; Srivastava, A.; Shalimar; Nayak, B.; Ct, R.K.; Surjit, M. Endoplasmic Reticulum Stress Induced Synthesis of a Novel Viral Factor Mediates Efficient Replication of Genotype-1 Hepatitis E Virus. PLoS Pathog. 2016, 12, e1005521. [Google Scholar] [CrossRef]
- Koonin, E.V.; Gorbalenya, A.E.; Purdy, M.A.; Rozanov, M.N.; Reyes, G.R.; Bradley, D.W. Computer-Assisted Assignment of Functional Domains in the Nonstructural Polyprotein of Hepatitis E Virus: Delineation of an Additional Group of Positive-Strand RNA Plant and Animal Viruses. Proc. Natl. Acad. Sci. USA 1992, 89, 8259–8263. [Google Scholar] [CrossRef]
- Agrawal, S.; Gupta, D.; Panda, S.K. The 3′ End of Hepatitis E Virus (HEV) Genome Binds Specifically to the Viral RNA-Dependent RNA Polymerase (RdRp). Virology 2001, 282, 87–101. [Google Scholar] [CrossRef]
- Cao, D.; Meng, X.-J. Molecular Biology and Replication of Hepatitis E Virus. Emerg. Microbes Infect. 2012, 1, e17. [Google Scholar] [CrossRef] [PubMed]
- Graff, J.; Torian, U.; Nguyen, H.; Emerson, S.U. A Bicistronic Subgenomic mRNA Encodes Both the ORF2 and ORF3 Proteins of Hepatitis E Virus. J. Virol. 2006, 80, 5919–5926. [Google Scholar] [CrossRef] [PubMed]
- Glitscher, M.; Hildt, E. Hepatitis E Virus Egress and beyond—The Manifold Roles of the Viral ORF3 Protein. Cell. Microbiol. 2021, 23, e13379. [Google Scholar] [CrossRef]
- Nagashima, S.; Takahashi, M.; Kobayashi, T.; Tanggis; Nishizawa, T.; Nishiyama, T.; Primadharsini, P.P.; Okamoto, H. Characterization of the Quasi-Enveloped Hepatitis E Virus Particles Released by the Cellular Exosomal Pathway. J. Virol. 2017, 91, e00822-17. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, M.; Yamada, K.; Hoshino, Y.; Takahashi, H.; Ichiyama, K.; Tanaka, T.; Okamoto, H. Monoclonal Antibodies Raised against the ORF3 Protein of Hepatitis E Virus (HEV) Can Capture HEV Particles in Culture Supernatant and Serum but Not Those in Feces. Arch. Virol. 2008, 153, 1703–1713. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Li, X.; Feng, Z. Role of Envelopment in the HEV Life Cycle. Viruses 2016, 8, 229. [Google Scholar] [CrossRef]
- Feng, Z.; Lemon, S.M. Peek-a-Boo: Membrane Hijacking and the Pathogenesis of Viral Hepatitis. Trends Microbiol. 2014, 22, 59–64. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Ambardekar, C.; Lu, Y.; Feng, Z. Distinct Entry Mechanisms for Nonenveloped and Quasi-Enveloped Hepatitis E Viruses. J. Virol. 2016, 90, 4232–4242. [Google Scholar] [CrossRef]
- Nelson, K.E.; Labrique, A.B.; Kmush, B.L. Epidemiology of Genotype 1 and 2 Hepatitis E Virus Infections. Cold Spring Harb. Perspect. Med. 2019, 9, a031732. [Google Scholar] [CrossRef]
- Huang, C.-C.; Nguyen, D.; Fernandez, J.; Yun, K.Y.; Fry, K.E.; Bradley, D.W.; Tam, A.W.; Reyes, G.R. Molecular Cloning and Sequencing of the Mexico Isolate of Hepatitis E Virus (HEV). Virology 1992, 191, 550–558. [Google Scholar] [CrossRef]
- Maila, H.T.; Bowyer, S.M.; Swanepoel, R. Identification of a New Strain of Hepatitis E Virus from an Outbreak in Namibia in 1995. J. Gen. Virol. 2004, 85, 89–95. [Google Scholar] [CrossRef]
- Nicand, E.; Armstrong, G.L.; Enouf, V.; Guthmann, J.P.; Guerin, J.-P.; Caron, M.; Nizou, J.Y.; Andraghetti, R. Genetic Heterogeneity of Hepatitis E Virus in Darfur, Sudan, and Neighboring Chad. J. Med. Virol. 2005, 77, 519–521. [Google Scholar] [CrossRef] [PubMed]
- Buisson, Y.; Grandadam, M.; Nicand, E.; Cheval, P.; van Cuyck-Gandre, H.; Innis, B.; Rehel, P.; Coursaget, P.; Teyssou, R.; Tsarev, S. Identification of a Novel Hepatitis E Virus in Nigeria. J. Gen. Virol. 2000, 81, 903–909. [Google Scholar] [CrossRef]
- Huang, R.; Nakazono, N.; Ishii, K.; Kawamata, O.; Kawaguchi, R.; Tsukada, Y. Existing Variations on the Gene Structure of Hepatitis E Virus Strains from Some Regions of China. J. Med. Virol. 1995, 47, 303–308. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.B.; Izopet, J.; Nicot, F.; Simmonds, P.; Jameel, S.; Meng, X.-J.; Norder, H.; Okamoto, H.; van der Poel, W.H.M.; Reuter, G.; et al. Update: Proposed Reference Sequences for Subtypes of Hepatitis E Virus (Species Orthohepevirus A). J. Gen. Virol. 2020, 101, 692–698. [Google Scholar] [CrossRef] [PubMed]
- Fanelli, A.; Tizzani, P.; Buonavoglia, D. A Systematic Review and Meta-Analysis of Hepatitis E Virus (HEV) in Wild Boars. Res. Vet. Sci. 2022, 142, 54–69. [Google Scholar] [CrossRef] [PubMed]
- Woo, P.C.Y.; Lau, S.K.P.; Teng, J.L.L.; Cao, K.-Y.; Wernery, U.; Schountz, T.; Chiu, T.H.; Tsang, A.K.L.; Wong, P.-C.; Wong, E.Y.M.; et al. New Hepatitis E Virus Genotype in Bactrian Camels, Xinjiang, China, 2013. Emerg. Infect. Dis. 2016, 22, 2219–2221. [Google Scholar] [CrossRef]
- Woo, P.C.Y.; Lau, S.K.P.; Teng, J.L.L.; Tsang, A.K.L.; Joseph, M.; Wong, E.Y.M.; Tang, Y.; Sivakumar, S.; Xie, J.; Bai, R.; et al. New Hepatitis E Virus Genotype in Camels, the Middle East. Emerg. Infect. Dis. 2014, 20, 1044–1048. [Google Scholar] [CrossRef]
- Pérez-Gracia, M.T.; García, M.; Suay, B.; Mateos-Lindemann, M.L. Current Knowledge on Hepatitis E. J. Clin. Transl. Hepatol. 2015, 3, 117–126. [Google Scholar] [CrossRef] [PubMed]
- Izopet, J.; Tremeaux, P.; Marion, O.; Migueres, M.; Capelli, N.; Chapuy-Regaud, S.; Mansuy, J.-M.; Abravanel, F.; Kamar, N.; Lhomme, S. Hepatitis E Virus Infections in Europe. J. Clin. Virol. 2019, 120, 20–26. [Google Scholar] [CrossRef]
- Zahmanova, G.; Takova, K.; Tonova, V.; Koynarski, T.; Lukov, L.L.; Minkov, I.; Pishmisheva, M.; Kotsev, S.; Tsachev, I.; Baymakova, M.; et al. The Re-Emergence of Hepatitis E Virus in Europe and Vaccine Development. Viruses 2023, 15, 1558. [Google Scholar] [CrossRef]
- Takahashi, M.; Okamoto, H. Features of Hepatitis E Virus Infection in Humans and Animals in Japan. Hepatol. Res. 2014, 44, 43–58. [Google Scholar] [CrossRef] [PubMed]
- Tei, S.; Kitajima, N.; Takahashi, K.; Mishiro, S. Zoonotic Transmission of Hepatitis E Virus from Deer to Human Beings. Lancet 2003, 362, 371–373. [Google Scholar] [CrossRef]
- Mrzljak, A.; Dinjar-Kujundzic, P.; Jemersic, L.; Prpic, J.; Barbic, L.; Savic, V.; Stevanovic, V.; Vilibic-Cavlek, T. Epidemiology of Hepatitis E in South-East Europe in the “One Health” Concept. World J. Gastroenterol. 2019, 25, 3168–3182. [Google Scholar] [CrossRef] [PubMed]
- Carpentier, A.; Chaussade, H.; Rigaud, E.; Rodriguez, J.; Berthault, C.; Boué, F.; Tognon, M.; Touzé, A.; Garcia-Bonnet, N.; Choutet, P.; et al. High Hepatitis E Virus Seroprevalence in Forestry Workers and in Wild Boars in France. J. Clin. Microbiol. 2012, 50, 2888–2893. [Google Scholar] [CrossRef] [PubMed]
- Ivanova, A.; Tefanova, V.; Reshetnjak, I.; Kuznetsova, T.; Geller, J.; Lundkvist, Å.; Janson, M.; Neare, K.; Velström, K.; Jokelainen, P.; et al. Hepatitis E Virus in Domestic Pigs, Wild Boars, Pig Farm Workers, and Hunters in Estonia. Food Environ. Virol. 2015, 7, 403–412. [Google Scholar] [CrossRef] [PubMed]
- Lange, H.; Øverbø, J.; Borgen, K.; Dudman, S.; Hoddevik, G.; Urdahl, A.M.; Vold, L.; Sjurseth, S.K. Hepatitis E in Norway: Seroprevalence in Humans and Swine. Epidemiol. Infect. 2017, 145, 181–186. [Google Scholar] [CrossRef] [PubMed]
- Salines, M.; Andraud, M.; Rose, N. From the Epidemiology of Hepatitis E Virus (HEV) within the Swine Reservoir to Public Health Risk Mitigation Strategies: A Comprehensive Review. Vet. Res. 2017, 48, 31. [Google Scholar] [CrossRef] [PubMed]
- Martinelli, N.; Luppi, A.; Cordioli, P.; Lombardi, G.; Lavazza, A. Prevalence of Hepatitis E Virus Antibodies in Pigs in Northern Italy. Infect. Ecol. Epidemiol. 2011, 1, 7331. [Google Scholar] [CrossRef]
- Seminati, C.; Mateu, E.; Peralta, B.; de Deus, N.; Martin, M. Distribution of Hepatitis E Virus Infection and Its Prevalence in Pigs on Commercial Farms in Spain. Vet. J. 2008, 175, 130–132. [Google Scholar] [CrossRef]
- Takova, K.; Koynarski, T.; Minkov, I.; Ivanova, Z.; Toneva, V.; Zahmanova, G. Increasing Hepatitis E Virus Seroprevalence in Domestic Pigs and Wild Boar in Bulgaria. Animals 2020, 10, 1521. [Google Scholar] [CrossRef]
- Tsachev, I.; Baymakova, M.; Marutsov, P.; Gospodinova, K.; Kundurzhiev, T.; Petrov, V.; Pepovich, R. Seroprevalence of Hepatitis E Virus Infection Among Wild Boars in Western Bulgaria. Vector-Borne Zoonotic Dis. 2021, 21, 441–445. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Si, F.; Jiang, C.; Li, T.; Jin, M. Molecular Detection of Hepatitis E Virus in Sheep from Southern Xinjiang, China. Virus Genes. 2015, 50, 410–417. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.; Li, Y.; Yu, W.; Jing, S.; Wang, J.; Long, F.; He, Z.; Yang, C.; Bi, Y.; Cao, W.; et al. Excretion of Infectious Hepatitis E Virus into Milk in Cows Imposes High Risks of Zoonosis. Hepatology 2016, 64, 350. [Google Scholar] [CrossRef] [PubMed]
- Rui, P.; Zhao, F.; Yan, S.; Wang, C.; Fu, Q.; Hao, J.; Zhou, X.; Zhong, H.; Tang, M.; Hui, W.; et al. Detection of Hepatitis E Virus Genotypes 3 and 4 in Donkeys in Northern China. Equine Vet. J. 2020, 52, 415–419. [Google Scholar] [CrossRef] [PubMed]
- García-Bocanegra, I.; Rivero, A.; Caballero-Gómez, J.; López-López, P.; Cano-Terriza, D.; Frías, M.; Jiménez-Ruiz, S.; Risalde, M.A.; Gómez-Villamandos, J.C.; Rivero-Juarez, A. Hepatitis E Virus Infection in Equines in Spain. Transbound. Emerg. Dis. 2019, 66, 66–71. [Google Scholar] [CrossRef] [PubMed]
- Doceul, V.; Bagdassarian, E.; Demange, A.; Pavio, N. Zoonotic Hepatitis E Virus: Classification, Animal Reservoirs and Transmission Routes. Viruses 2016, 8, 270. [Google Scholar] [CrossRef] [PubMed]
- Eiden, M.; Dähnert, L.; Spoerel, S.; Vina-Rodriguez, A.; Schröder, R.; Conraths, F.J.; Groschup, M.H. Spatial-Temporal Dynamics of Hepatitis E Virus Infection in Foxes (Vulpes vulpes) in Federal State of Brandenburg, Germany, 1993–2012. Front. Microbiol. 2020, 11, 115. [Google Scholar] [CrossRef] [PubMed]
- Caballero-Gómez, J.; Rivero-Juarez, A.; Jurado-Tarifa, E.; Jiménez-Martín, D.; Jiménez-Ruiz, E.; Castro-Scholten, S.; Ulrich, R.G.; López-López, P.; Rivero, A.; García-Bocanegra, I. Serological and Molecular Survey of Hepatitis E Virus in Cats and Dogs in Spain. Transbound. Emerg. Dis. 2022, 69, 240–248. [Google Scholar] [CrossRef]
- Li, S.; Liu, M.; Cong, J.; Zhou, Y.; Miao, Z. Detection and Characterization of Hepatitis E Virus in Goats at Slaughterhouse in Tai’an Region, China. BioMed Res. Int. 2017, 2017, e3723650. [Google Scholar] [CrossRef]
- Tsachev, I.; Gospodinova, K.; Pepovich, R.; Takova, K.; Kundurzhiev, T.; Zahmanova, G.; Kaneva, K.; Baymakova, M. First Insight into the Seroepidemiology of Hepatitis E Virus (HEV) in Dogs, Cats, Horses, Cattle, Sheep, and Goats from Bulgaria. Viruses 2023, 15, 1594. [Google Scholar] [CrossRef]
- Mesquita, J.R.; Istrate, C.; Santos-Ferreira, N.L.; Ferreira, A.S.; Abreu-Silva, J.; Veiga, J.; van der Poel, W.H.M.; Nascimento, M.S.J. Short Communication: Detection and Molecular Characterization of Hepatitis E Virus in Domestic Animals of São Tomé and Príncipe. Trop. Anim. Health Prod. 2019, 51, 481–485. [Google Scholar] [CrossRef] [PubMed]
- Long, F.; Yu, W.; Yang, C.; Wang, J.; Li, Y.; Li, Y.; Huang, F. High Prevalence of Hepatitis E Virus Infection in Goats. J. Med. Virol. 2017, 89, 1981–1987. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Pan, Y.; Baloch, A.R.; Tian, L.; Wang, M.; Na, W.; Ding, L.; Zeng, Q. Hepatitis E Virus Genotype 4 in Yak, Northwestern China. Emerg. Infect. Dis. J.-CDC 2014, 20, 12. [Google Scholar] [CrossRef]
- Wolfe, B.A. Bovidae (Except Sheep and Goats) and Antilocapridae. Fowler’s Zoo. Wild Anim. Med. 2015, 8, 626–645. [Google Scholar] [CrossRef]
- Peralta, B.; Casas, M.; de Deus, N.; Martín, M.; Ortuño, A.; Pérez-Martín, E.; Pina, S.; Mateu, E. Anti-HEV Antibodies in Domestic Animal Species and Rodents from Spain Using a Genotype 3-Based ELISA. Vet. Microbiol. 2009, 137, 66–73. [Google Scholar] [CrossRef] [PubMed]
- Yugo, D.M.; Cossaboom, C.M.; Heffron, C.L.; Huang, Y.-W.; Kenney, S.P.; Woolums, A.R.; Hurley, D.J.; Opriessnig, T.; Li, L.; Delwart, E.; et al. Evidence for an Unknown Agent Antigenically Related to the Hepatitis E Virus in Dairy Cows in the United States. J. Med. Virol. 2019, 91, 677–686. [Google Scholar] [CrossRef] [PubMed]
- Vitral, C.L.; Pinto, M.A.; Lewis-Ximenez, L.L.; Khudyakov, Y.E.; dos Santos, D.R.; Gaspar, A.M.C. Serological Evidence of Hepatitis E Virus Infection in Different Animal Species from the Southeast of Brazil. Mem. Inst. Oswaldo Cruz 2005, 100, 117–122. [Google Scholar] [CrossRef] [PubMed]
- Arankalle, V.A.; Joshi, M.V.; Kulkarni, A.M.; Gandhe, S.S.; Chobe, L.P.; Rautmare, S.S.; Mishra, A.C.; Padbidri, V.S. Prevalence of Anti-Hepatitis E Virus Antibodies in Different Indian Animal Species. J. Viral Hepat. 2001, 8, 223–227. [Google Scholar] [CrossRef]
- Fu, H.; Li, L.; Zhu, Y.; Wang, L.; Geng, J.; Chang, Y.; Xue, C.; Du, G.; Li, Y.; Zhuang, H. Hepatitis E Virus Infection among Animals and Humans in Xinjiang, China: Possibility of Swine to Human Transmission of Sporadic Hepatitis E in an Endemic Area. Am. J. Trop. Med. Hyg. 2010, 82, 961–966. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Sun, J.; Liu, M.; Xia, L.; Zhao, C.; Harrison, T.J.; Wang, Y. Seroepidemiology and Genetic Characterization of Hepatitis E Virus in the Northeast of China. Infect. Genet. Evol. 2009, 9, 554–561. [Google Scholar] [CrossRef]
- Geng, Y.; Wang, C.; Zhao, C.; Yu, X.; Harrison, T.J.; Tian, K.; Wang, Y. Serological Prevalence of Hepatitis E Virus in Domestic Animals and Diversity of Genotype 4 Hepatitis E Virus in China. Vector Borne Zoonotic Dis. 2010, 10, 765–770. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.; Wang, L.; Geng, J.; Zhu, Y.; Fu, H.; Ren, F.; Li, L.; Wang, X.; Zhuang, H. Zoonotic Risk of Hepatitis E Virus (HEV): A Study of HEV Infection in Animals and Humans in Suburbs of Beijing. Hepatol. Res. 2009, 39, 1153–1158. [Google Scholar] [CrossRef] [PubMed]
- Tonbak, F.; Atasever, M. Determination of Hepatitis E Virus in Sheep and Cattle by Serological and Molecular Methods DNA Sequences Analysis. Acta Vet. Eurasia 2022, 48, 94–100. [Google Scholar] [CrossRef]
- Obaidat, M.M.; Roess, A.A. Individual Animal and Herd Level Seroprevalence and Risk Factors of Hepatitis E in Ruminants in Jordan. Infect. Genet. Evol. 2020, 81, 104276. [Google Scholar] [CrossRef]
- El-Tras, W.F.; Tayel, A.A.; El-Kady, N.N. Seroprevalence of Hepatitis E Virus in Humans and Geographically Matched Food Animals in Egypt. Zoonoses Public Health 2013, 60, 244–251. [Google Scholar] [CrossRef]
- Tialla, D.; Cissé, A.; Ouédraogo, G.A.; Hübschen, J.M.; Tarnagda, Z.; Snoeck, C.J. Prevalence of Hepatitis E Virus Antibodies in Cattle in Burkina Faso Associated with Swine Mixed Farming. J. Vet. Sci. 2022, 23, e33. [Google Scholar] [CrossRef] [PubMed]
- Ouoba, J.B.; Traore, K.A.; Rouamba, H.; Setondji, K.V.-M.; Minoungou, G.L.; Ouoba, B.L.; Ouedraogo, A.; Moctar, S.; M’Bengue, A.K.; Kakou, S.N.; et al. Prevalence of Anti-Hepatitis E Virus Antibodies in Domestic Animal from Three Representative Provinces of Burkina Faso. Vet. Anim. Sci. 2019, 7, 100059. [Google Scholar] [CrossRef]
- Song, Y.-J.; Jeong, H.-J.; Kim, Y.-J.; Lee, S.-W.; Lee, J.-B.; Park, S.-Y.; Song, C.-S.; Park, H.-M.; Choi, I.-S. Analysis of Complete Genome Sequences of Swine Hepatitis E Virus and Possible Risk Factors for Transmission of HEV to Humans in Korea. J. Med. Virol. 2010, 82, 583–591. [Google Scholar] [CrossRef]
- Junaid, S.A.; Agina, S.E.; Jaiye, K. Seroprevalence of Hepatitis E Virus among Domestic Animals in Plateau State–Nigeria. Microbiol. Res. J. Int. 2014, 4, 924–934. [Google Scholar] [CrossRef]
- Antia, R.E.; Adekola, A.A.; Jubril, A.J.; Ohore, O.G.; Emikpe, B.O. Hepatitis E Virus Infection Seroprevalence and the Associated Risk Factors in Animals Raised in Ibadan, Nigeria. J. Immunoass. Immunochem. 2018, 39, 509–520. [Google Scholar] [CrossRef]
- Sarchese, V.; Di Profio, F.; Melegari, I.; Palombieri, A.; Sanchez, S.B.; Arbuatti, A.; Ciuffetelli, M.; Marsilio, F.; Martella, V.; Di Martino, B. Hepatitis E Virus in Sheep in Italy. Transbound. Emerg. Dis. 2019, 66, 1120–1125. [Google Scholar] [CrossRef] [PubMed]
- Palombieri, A.; Robetto, S.; Di Profio, F.; Sarchese, V.; Fruci, P.; Bona, M.C.; Ru, G.; Orusa, R.; Marsilio, F.; Martella, V.; et al. Surveillance Study of Hepatitis E Virus (HEV) in Domestic and Wild Ruminants in Northwestern Italy. Animals 2020, 10, 2351. [Google Scholar] [CrossRef] [PubMed]
- Mesquita, J.R.; Santos-Ferreira, N.; Ferreira, A.S.; Albuquerque, C.; Nóbrega, C.; Esteves, F.; Cruz, R.; Vala, H.; Nascimento, M.S.J. Increased Risk of Hepatitis E Virus Infection in Workers Occupationally Exposed to Sheep. Transbound. Emerg. Dis. 2020, 67, 1918–1921. [Google Scholar] [CrossRef] [PubMed]
- Caballero-Gómez, J.; García-Bocanegra, I.; Jiménez-Martín, D.; Cano-Terriza, D.; Risalde, M.A.; López-López, P.; Jiménez-Ruiz, S.; Rivero, A.; Rivero-Juarez, A. Epidemiological Survey and Risk Factors Associated with Hepatitis E Virus in Small Ruminants in Southern Spain. Zoonoses Public Health 2022, 69, 387–393. [Google Scholar] [CrossRef] [PubMed]
- Geng, J.; Wang, L.; Wang, X.; Fu, H.; Bu, Q.; Liu, P.; Zhu, Y.; Wang, M.; Sui, Y.; Zhuang, H. Potential Risk of Zoonotic Transmission from Young Swine to Human: Seroepidemiological and Genetic Characterization of Hepatitis E Virus in Human and Various Animals in Beijing, China. J. Viral Hepat. 2011, 18, e583–e590. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Kang, Q.; Bai, W.; Bai, Z. Seroepidemiological survey of sheep hepatitis E virus infection in Aksu region of Xinjiang Autonomous. Bing. Du. Xue Bao 2010, 26, 234–237. [Google Scholar] [PubMed]
- Shuaibu, A.B.; Alkali, B.R.; Abubakar, M.B.; Daneji, A.I.; Shuaibu, S.A.; Bello, A.I.; Abubaka, F.; Bello, M. Prevalence of Hepatitis E Virus (HEV) Antibodies in Sheep from Sokoto State. J. Adv. Microbiol. 2016, 1, 1–6. [Google Scholar] [CrossRef]
- Zhang, W.; Shen, Q.; Mou, J.; Gong, G.; Yang, Z.; Cui, L.; Zhu, J.; Ju, G.; Hua, X. Hepatitis E Virus Infection among Domestic Animals in Eastern China. Zoonoses Public Health 2008, 55, 291–298. [Google Scholar] [CrossRef] [PubMed]
- Shukla, P.; Chauhan, U.K.; Naik, S.; Anderson, D.; Aggarwal, R. Hepatitis E Virus Infection among Animals in Northern India: An Unlikely Source of Human Disease. J. Viral Hepat. 2007, 14, 310–317. [Google Scholar] [CrossRef]
- Sanford, B.J.; Emerson, S.U.; Purcell, R.H.; Engle, R.E.; Dryman, B.A.; Cecere, T.E.; Buechner-Maxwell, V.; Sponenberg, D.P.; Meng, X.J. Serological Evidence for a Hepatitis E Virus-Related Agent in Goats in the United States. Transbound. Emerg. Dis. 2013, 60, 538–545. [Google Scholar] [CrossRef]
- Deng Yu, D.Y. Serological Epidemiology of Goat Hepatitis E in Panxi Area of Sichuan Province. Zhongguo Yufang Shouyi Xuebao/Chin. J. Prev. Vet. Med. 2014, 36, 805–806. [Google Scholar]
- Wang, Y.-C.; Zhang, H.-Y.; Xia, N.-S.; Peng, G.; Lan, H.-Y.; Zhuang, H.; Zhu, Y.-H.; Li, S.-W.; Tian, K.-G.; Gu, W.-J.; et al. Prevalence, Isolation, and Partial Sequence Analysis of Hepatitis E Virus from Domestic Animals in China. J. Med. Virol. 2002, 67, 516–521. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, M.; Nishizono, A.; Kawakami, M.; Fukui, E.; Isogai, E.; Matsuoka, H.; Yamamoto, S.; Mizuo, H.; Nagashima, S.; Murata, K.; et al. Identification of Hepatitis E Virus in Wild Sika Deer in Japan. Virus Res. 2022, 308, 198645. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Shen, Q.; Mou, J.; Yang, Z.B.; Yuan, C.L.; Cui, L.; Zhu, J.G.; Hua, X.G.; Xu, C.M.; Hu, J. Cross-Species Infection of Hepatitis E Virus in a Zoo-like Location, Including Birds. Epidemiol. Infect. 2008, 136, 1020–1026. [Google Scholar] [CrossRef] [PubMed]
- Fonti, N.; Pacini, M.I.; Forzan, M.; Parisi, F.; Periccioli, M.; Mazzei, M.; Poli, A. Molecular and Pathological Detection of Hepatitis E Virus in Roe Deer (Capreolus capreolus) and Fallow Deer (Dama dama) in Central Italy. Vet. Sci. 2022, 9, 100. [Google Scholar] [CrossRef]
- Lin, J.; Norder, H.; Uhlhorn, H.; Belák, S.; Widén, F. Novel Hepatitis E like Virus Found in Swedish Moose. J. Gen. Virol. 2014, 95, 557–570. [Google Scholar] [CrossRef]
- Santos-Silva, S.; López-López, P.; Gonçalves, H.M.R.; Rivero-Juarez, A.; Van der Poel, W.H.M.; Nascimento, M.S.J.; Mesquita, J.R. A Systematic Review and Meta-Analysis on Hepatitis E Virus Detection in Farmed Ruminants. Pathogens 2023, 12, 550. [Google Scholar] [CrossRef] [PubMed]
- Demirci, M.; Yiğin, A.; Ünlü, Ö.; Altun, S.K. Detection of HEV RNA Amounts and Genotypes in Raw Milks Obtained from Different Animals. Mikrobiyoloji Bul. 2019, 53, 43–52. [Google Scholar] [CrossRef]
- Trojnar, E.; Kästner, B.; Johne, R. No Evidence of Hepatitis E Virus Infection in Farmed Deer in Germany. Food Environ. Virol. 2020, 12, 81–83. [Google Scholar] [CrossRef]
- Schlosser, J.; Dähnert, L.; Dremsek, P.; Tauscher, K.; Fast, C.; Ziegler, U.; Gröner, A.; Ulrich, R.G.; Groschup, M.H.; Eiden, M. Different Outcomes of Experimental Hepatitis E Virus Infection in Diverse Mouse Strains, Wistar Rats, and Rabbits. Viruses 2019, 11, 1. [Google Scholar] [CrossRef]
- Di Martino, B.; Di Profio, F.; Melegari, I.; Sarchese, V.; Robetto, S.; Marsilio, F.; Martella, V. Detection of Hepatitis E Virus (HEV) in Goats. Virus Res. 2016, 225, 69–72. [Google Scholar] [CrossRef] [PubMed]
- Batmagnai, E.; Boldbaatar, B.; Sodbayasgalan, A.; Kato-Mori, Y.; Hagiwara, K. Hepatitis E Virus (HEV) Spreads from Pigs and Sheep in Mongolia. Animals 2023, 13, 891. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Ma, X. Detection and sequences analysis of sheep hepatitis E virus RNA in Xinjiang autonomous region. Wei Sheng Wu Xue Bao 2010, 50, 937–941. [Google Scholar] [PubMed]
- Hu, G.-D.; Ma, X. Detection and sequences analysis of bovine hepatitis E virus RNA in Xinjiang Autonomous Region. Bing Du Xue Bao 2010, 26, 27–32. [Google Scholar]
- Go, H.-J.; Park, B.-J.; Ahn, H.-S.; Lyoo, E.-L.; Kim, D.-H.; Lee, J.-B.; Park, S.-Y.; Song, C.-S.; Lee, S.-W.; Choi, I.-S. Identification of Hepatitis E Virus in Bovine and Porcine Raw Livers. J. Microbiol. Biotechnol. 2019, 29, 2022–2025. [Google Scholar] [CrossRef] [PubMed]
- Rahmani, F.; Hmaied, F.; Matei, I.; Chirila, F.; Fit, N.; Yahya, M.; Jebri, S.; Amairia, S.; Hamdi, M. Occurrence of Staphylococcus Spp. and Investigation of Fecal and Animal Viral Contaminations in Livestock, River Water, and Sewage from Tunisia and Romania. Environ. Monit. Assess. 2020, 192, 206. [Google Scholar] [CrossRef] [PubMed]
- Yan, B.; Zhang, L.; Gong, L.; Lv, J.; Feng, Y.; Liu, J.; Song, L.; Xu, Q.; Jiang, M.; Xu, A. Hepatitis E Virus in Yellow Cattle, Shandong, Eastern China. Emerg. Infect. Dis. J.-CDC 2016, 22, 2211. [Google Scholar] [CrossRef] [PubMed]
- Uema, M.; Yonemitsu, K.; Sasaki, Y.; Asakura, H. Detection of Hepatitis E Virus RNA from Pig Bile Collected at a Slaughterhouse in Japan. AIMS Microbiol. 2022, 8, 566–574. [Google Scholar] [CrossRef] [PubMed]
- Boxman, I.L.A.; Verhoef, L.; Dop, P.Y.; Vennema, H.; Dirks, R.A.M.; Opsteegh, M. High Prevalence of Acute Hepatitis E Virus Infection in Pigs in Dutch Slaughterhouses. Int. J. Food Microbiol. 2022, 379, 109830. [Google Scholar] [CrossRef]
- García, N.; Hernández, M.; Gutierrez-Boada, M.; Valero, A.; Navarro, A.; Muñoz-Chimeno, M.; Fernández-Manzano, A.; Escobar, F.M.; Martínez, I.; Bárcena, C.; et al. Occurrence of Hepatitis E Virus in Pigs and Pork Cuts and Organs at the Time of Slaughter, Spain, 2017. Front. Microbiol. 2019, 10, 2990. [Google Scholar] [CrossRef]
- Pellerin, M.; Trabucco, B.; Capai, L.; Laval, M.; Maestrini, O.; Jori, F.; Falchi, A.; Doceul, V.; Charrier, F.; Casabianca, F.; et al. Low Prevalence of Hepatitis E Virus in the Liver of Corsican Pigs Slaughtered after 12 Months despite High Antibody Seroprevalence. Transbound. Emerg. Dis. 2022, 69, e2706–e2718. [Google Scholar] [CrossRef] [PubMed]
- Milojevic, L.; Velebit, B.; Teodorovic, V.; Kirbis, A.; Petrovic, T.; Karabasil, N.; Dimitrijevic, M. Screening and Molecular Characterization of Hepatitis E Virus in Slaughter Pigs in Serbia. Food Environ. Virol. 2019, 11, 410–419. [Google Scholar] [CrossRef] [PubMed]
- El-Mokhtar, M.A.; Elkhawaga, A.A.; Sayed, I.M. Assessment of Hepatitis E Virus (HEV) in the Edible Goat Products Pointed out a Risk for Human Infection in Upper Egypt. Int. J. Food Microbiol. 2020, 330, 108784. [Google Scholar] [CrossRef] [PubMed]
- Dziedzinska, R.; Krzyzankova, M.; Bena, M.; Vasickova, P. Evidence of Hepatitis E Virus in Goat and Sheep Milk. Viruses 2020, 12, 1429. [Google Scholar] [CrossRef] [PubMed]
- Tritz, S.E.; Khounvisith, V.; Pommasichan, S.; Ninnasopha, K.; Keosengthong, A.; Phoutana, V.; Camoin, M.; Hübschen, J.M.; Black, A.P.; Muller, C.P.; et al. Evidence of Increased Hepatitis E Virus Exposure in Lao Villagers with Contact to Ruminants. Zoonoses Public Health 2018, 65, 690–701. [Google Scholar] [CrossRef] [PubMed]
- Shao, Z.; Li, J.; Zheng, Y.; Zhang, J.; Ma, Y.; Ma, W.; Jiang, Q.; Dang, R. Epidemiological Screening for Hepatitis E Virus in Bile Specimens from Livestock in Northwest China. J. Clin. Microbiol. 2009, 47, 814–816. [Google Scholar] [CrossRef] [PubMed]
- Wei, D.; Zhao, Y.; Jia, Y.; Hao, X.; Situ, J.; Yu, W.; Huang, F.; Jiang, H. Hepatitis E Virus Infection in Buffaloes in South China. Arq. Bras. Med. Vet. Zootec. 2020, 72, 1122–1126. [Google Scholar] [CrossRef]
- Sayed, I.M.; Elkhawaga, A.A.; El-Mokhtar, M.A. Circulation of Hepatitis E Virus (HEV) and/or HEV-like Agent in Non-Mixed Dairy Farms Could Represent a Potential Source of Infection for Egyptian People. Int. J. Food Microbiol. 2020, 317, 108479. [Google Scholar] [CrossRef] [PubMed]
- Bastos, C.; Eisen, A.K.A.; Demoliner, M.; Heldt, F.H.; Filippi, M.; de Abreu Góes Pereira, V.M.; Teixeira, T.A.M.; Roth, L.O.; Gularte, J.S.; Spilki, F.R. Hepatitis E Virus Genotype 3 in Bovine Livers Slaughtered in the State of Rio Grande Do Sul, Brazil. Braz. J. Microbiol. 2022, 53, 1115–1120. [Google Scholar] [CrossRef]
- Santos-Silva, S.; Gonçalves, H.M.R.; Rivero-Juarez, A.; Van der Poel, W.H.M.; Nascimento, M.S.J.; Mesquita, J.R. Detection of Hepatitis E Virus in Milk: Current Evidence for Viral Excretion in a Wide Range of Mammalian Hosts. Transbound. Emerg. Dis. 2022, 69, 3173–3180. [Google Scholar] [CrossRef]
- Baechlein, C.; Becher, P. No Evidence for Zoonotic Hepatitis E Virus Infection through Dairy Milk in Germany. Hepatology 2017, 65, 394. [Google Scholar] [CrossRef] [PubMed]
- Vercouter, A.-S.; Sayed, I.M.; Lipkens, Z.; De Bleecker, K.; De Vliegher, S.; Colman, R.; Koppelman, M.; Supré, K.; Meuleman, P. Absence of Zoonotic Hepatitis E Virus Infection in Flemish Dairy Cows. Int. J. Food Microbiol. 2018, 281, 54–59. [Google Scholar] [CrossRef] [PubMed]
- Sayed, I.M.; Hammam, A.R.A.; Elfaruk, M.S.; Alsaleem, K.A.; Gaber, M.A.; Ezzat, A.A.; Salama, E.H.; Elkhawaga, A.A.; El-Mokhtar, M.A. Enhancement of the Molecular and Serological Assessment of Hepatitis E Virus in Milk Samples. Microorganisms 2020, 8, 1231. [Google Scholar] [CrossRef] [PubMed]
- Rivero-Juarez, A.; Frias, M.; Rodriguez-Cano, D.; Cuenca-López, F.; Rivero, A. Isolation of Hepatitis E Virus From Breast Milk During Acute Infection. Clin. Infect. Dis. 2016, 62, 1464. [Google Scholar] [CrossRef] [PubMed]
- Feagins, A.R.; Opriessnig, T.; Guenette, D.K.; Halbur, P.G.; Meng, X.J. Inactivation of Infectious Hepatitis E Virus Present in Commercial Pig Livers Sold in Local Grocery Stores in the United States. Int. J. Food Microbiol. 2008, 123, 32–37. [Google Scholar] [CrossRef] [PubMed]
- Johne, R.; Trojnar, E.; Filter, M.; Hofmann, J. Thermal Stability of Hepatitis E Virus as Estimated by a Cell Culture Method. Appl. Env. Microbiol. 2016, 82, 4225–4231. [Google Scholar] [CrossRef]
- Imagawa, T.; Sugiyama, R.; Shiota, T.; Li, T.-C.; Yoshizaki, S.; Wakita, T.; Ishii, K. Evaluation of Heating Conditions for Inactivation of Hepatitis E Virus Genotypes 3 and 4. J. Food Prot. 2018, 81, 947–952. [Google Scholar] [CrossRef] [PubMed]
- Kamar, N.; Pischke, S. Acute and Persistent Hepatitis E Virus Genotype 3 and 4 Infection: Clinical Features, Pathogenesis, and Treatment. Cold Spring Harb. Perspect. Med. 2019, 9, a031872. [Google Scholar] [CrossRef] [PubMed]
- Nasheri, N.; Doctor, T.; Chen, A.; Harlow, J.; Gill, A. Evaluation of High-Pressure Processing in Inactivation of the Hepatitis E Virus. Front. Microbiol. 2020, 11, 461. [Google Scholar] [CrossRef]
- Bouquet, P.; Alexandre, V.; De Lamballerie, M.; Ley, D.; Lesage, J.; Goffard, A.; Cocquerel, L. Effect of High Hydrostatic Pressure Processing and Holder Pasteurization of Human Milk on Inactivation of Human Coronavirus 229E and Hepatitis E Virus. Viruses 2023, 15, 1571. [Google Scholar] [CrossRef]
- Usmanov, R.K.; Balaian, M.S.; Dvoĭnikova, O.V.; Alymbaeva, D.B.; Zamiatina, N.A.; Kazachkov, I.A.; Belov, V.I. An experimental infection in lambs by the hepatitis E virus. Vopr. Virusol. 1994, 39, 165–168. [Google Scholar] [PubMed]
- Qian, Z.; Hao, X.; Xia, Y.; Yu, W.; Huang, F. Rat Hepatitis E Virus Is a Potential Zoonotic Pathogen to Humans. J. Hepatol. 2022, 77, 868–870. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Kitajima, N.; Abe, N.; Mishiro, S. Complete or Near-Complete Nucleotide Sequences of Hepatitis E Virus Genome Recovered from a Wild Boar, a Deer, and Four Patients Who Ate the Deer. Virology 2004, 330, 501–505. [Google Scholar] [CrossRef] [PubMed]
| Animal | Sample Type | RNA Detection | Genotype (gt) | Reference |
|---|---|---|---|---|
| Goat | Faeces | 9.2% (11/119) | P. balayani gt3 | [101] |
| Faeces Serum | 74.1% (40/54) | P. balayani gt4 | [62] | |
| 60.0% (12/20) | ||||
| 53.5% (15/28) | ||||
| Liver | 4.0% (2/50) | P. balayani gt4 | [59] | |
| Liver | ||||
| Sheep | Faeces | 10.4% (20/192) | P. balayani gt3 | [81] |
| Serum | 1.6% (3/192) | |||
| Liver | 5.3% (4/75) | P. balayani gt4 | [52] | |
| Liver | 5% (3/60) | No data | [102] | |
| Faeces | 2% (4/200) | |||
| Faeces | 11.1% (6/54) | P. balayani gt4 | [103] | |
| Cow | Faeces | 37.14% (52/140) | P. balayani gt4h | [53] |
| Faeces | 7. 9% (8/91) | P. balayani gt4 | [104] | |
| Liver | 1.0% (1/100) | P. balayani gt4 | [105] | |
| Faeces | 2.04% (1/45) | No data | [106] | |
| Faeces | 14.29% (1/7) | P. balayani 3f | [61] | |
| Yellow cattle (Bos taurus) | Serum | 3% (8/254) | P. balayani gt4d | [107] |
| Yak (Bos grunniens) | Faeces | 3.26% (3/92) | P. balayani gt4/3 | [63] |
| Animal | Sample Type and Location of Sampling | RNA Detection | Viral Load IU/mL | Genotype (gt) | References |
|---|---|---|---|---|---|
| Cow | Bulk milk from industrial farms, Germany | 0% (0/400) | - | No data | [121] |
| Cow | Bulk Milk from industrial farms, Belgium and the Netherlands | 0% (0/504) | - | No data | [122] |
| Cow | Milk from traditional small farms, Turkey | 29.2% (14/48) | - | P. balayani gt1, gt3, gt4 | [98] |
| Cow | Milk from traditional farming animals, China | 37% (52/140) | - | P. balayani gt4h | [53] |
| Cow | Milk from nonmixed dairy farms, Egypt | 0.2% (1/480) | 103 IU/mL | P. balayani gt3a | [118,123] |
| Buffalo | Milk from traditional farming animals, China | 7.5% (3/40) | - | P. balayani gt4 | [117] |
| Goat | Milk from mixed dairy farms, China | 100% (4/4) | 104 to 105 IU/mL | P. balayani gt4h | [62] |
| Goat | Milk from traditional farming animals in villages, Egypt | 0.7% (2/280) HEV Ag (1.8%) | - | P. balayani gt3 | [113] |
| Goat | Milk from traditional farming animals, Turkey | 18.46% (12/65) | P. balayani gt1, gt3, gt4 | [98] | |
| Goat and Sheep | 938 sheep and 2674 goat milk samples were pooled into 290 samples, Czech Republic | 2.9% (8/290) | 101 to 103 IU/mL | No data | [114] |
| Sheep | Milk from mixed dairy farms, Turkey | 12.3% (8/65) | - | P. balayani gt1, gt3, gt4 | [98] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zahmanova, G.; Takova, K.; Lukov, G.L.; Andonov, A. Hepatitis E Virus in Domestic Ruminants and Virus Excretion in Milk—A Potential Source of Zoonotic HEV Infection. Viruses 2024, 16, 684. https://doi.org/10.3390/v16050684
Zahmanova G, Takova K, Lukov GL, Andonov A. Hepatitis E Virus in Domestic Ruminants and Virus Excretion in Milk—A Potential Source of Zoonotic HEV Infection. Viruses. 2024; 16(5):684. https://doi.org/10.3390/v16050684
Chicago/Turabian StyleZahmanova, Gergana, Katerina Takova, Georgi L. Lukov, and Anton Andonov. 2024. "Hepatitis E Virus in Domestic Ruminants and Virus Excretion in Milk—A Potential Source of Zoonotic HEV Infection" Viruses 16, no. 5: 684. https://doi.org/10.3390/v16050684






