Human Cytomegalovirus Encoded Homologs of Cytokines, Chemokines and their Receptors: Roles in Immunomodulation
Abstract
:1. Introduction
| Gene name | Homology | Function(s) | Reference(s) |
|---|---|---|---|
| UL21.5 | Soluble chemokine receptor | Binds CCL5 preventing host cell signaling | 109 |
| US27 | Chemokine receptor | Role in extracellular spread of virus | 102 |
| US28 | Chemokine receptor | Potential oncogene | 96-99 |
| Promotes chemotaxis | 77 | ||
| Potential chemokine sink | 91-93 | ||
| UL33 | Chemokine receptor | Modulates CXCR4 and CCL5 activity | 108 |
| Modulates pUS28 activity | 104 | ||
| UL78 | Chemokine receptor | Modulates CXCR4 and CCL5 activity | 108 |
| Modulates pUS28 activity | 104 | ||
| UL111A(cmvIL-10) | Cytokine | Inhibits myeloid cell functions | 10-17 |
| Stimulates B cell proliferation | 18 | ||
| UL111A(LAcmvIL-10) | Cytokine | Inhibits MHC class II expression | 22 |
| UL128 | Chemokine | Promotes PBMC migration | 50 |
| UL144 | Cytokine receptor | Inhibits T-cell proliferation via BTLA-4 | 38 |
| Induces CCL22 via NF-KB | 37, 41 | ||
| UL146 | Chemokine | Promotes neutrophil chemotaxis | 44 |
| UL147 | Chemokine | Unknown function |
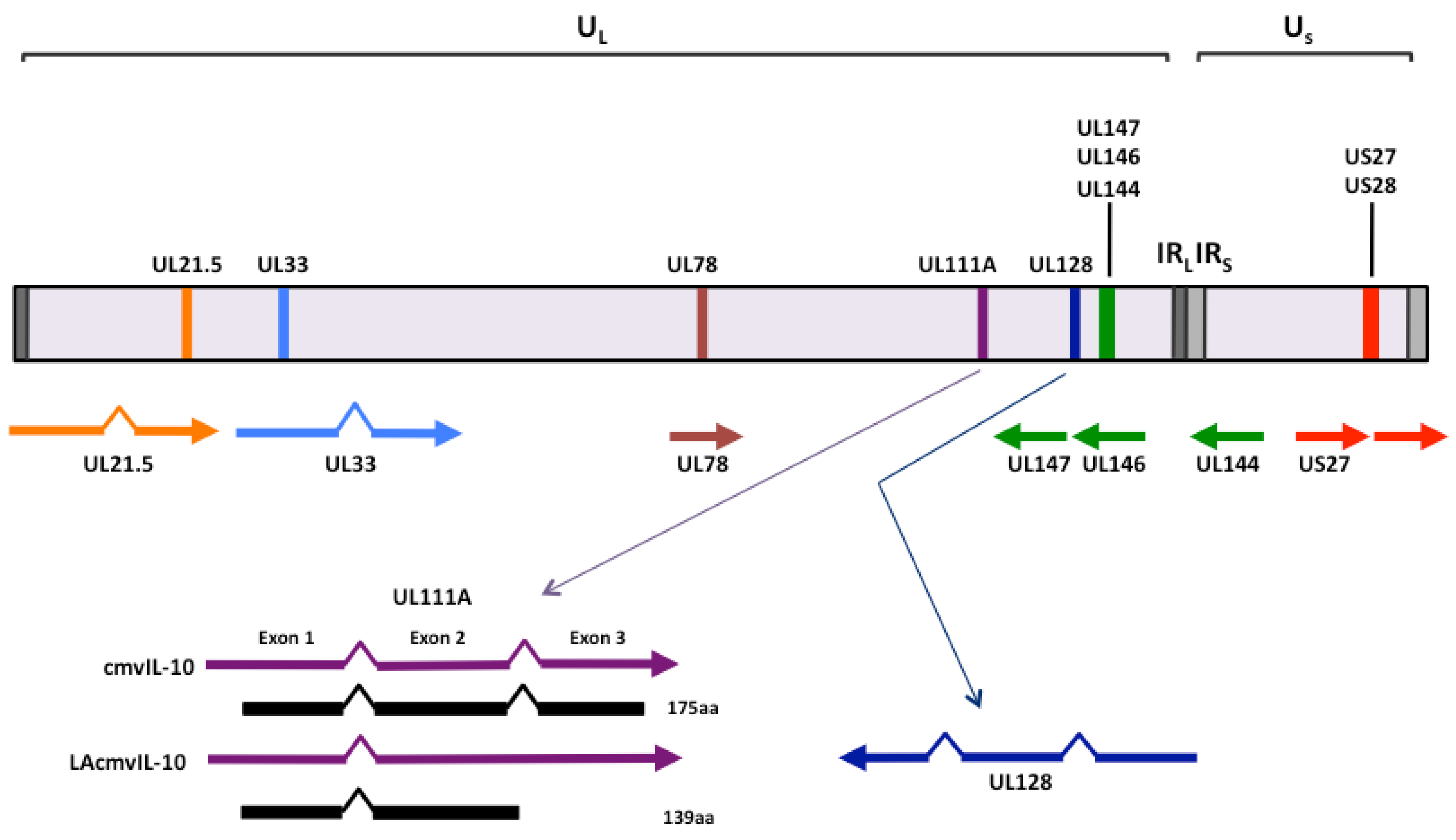
2. Cytokine homologs encoded by human cytomegalovirus UL111A
2.1 cmvIL-10 and LAcmvIL-10
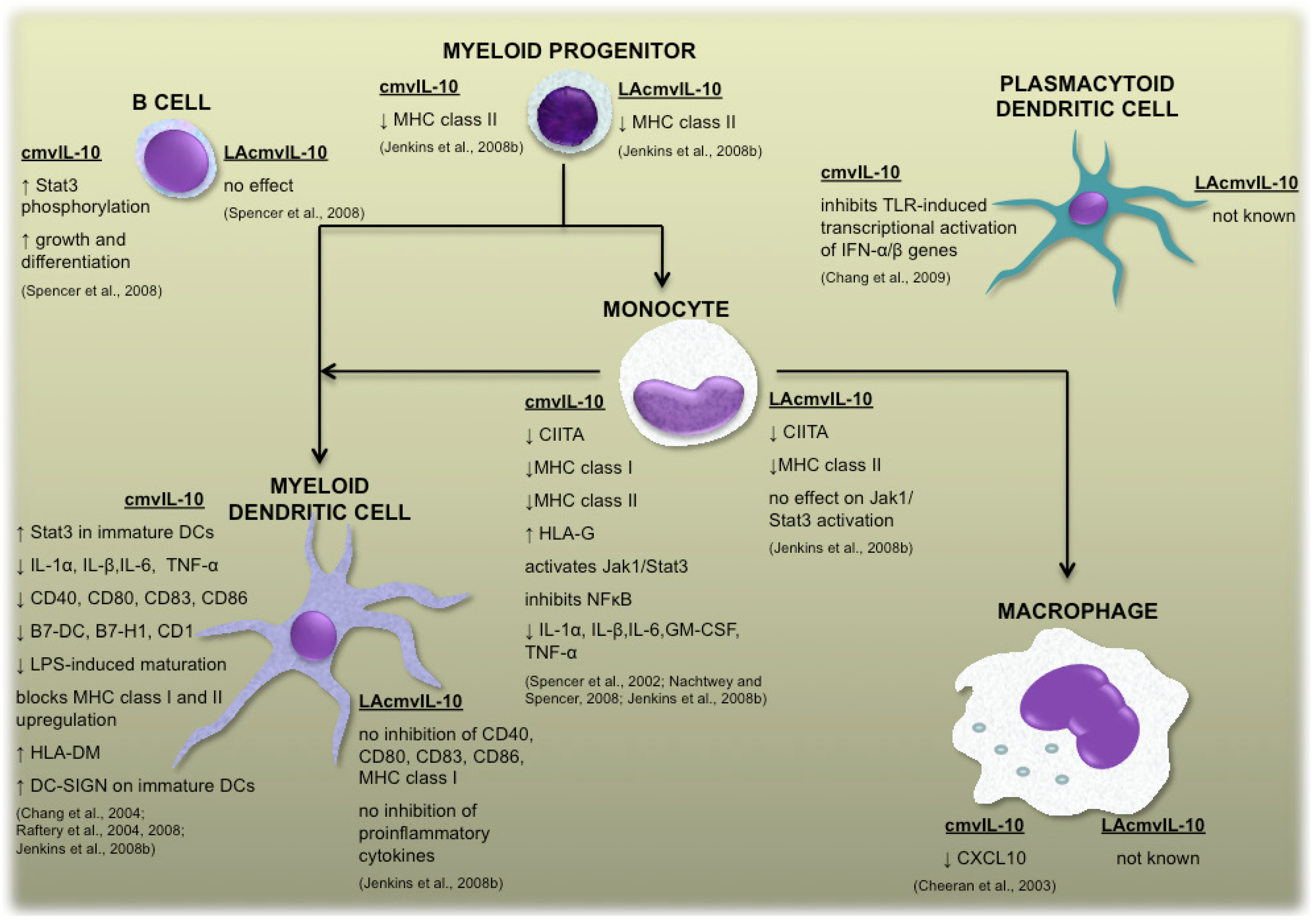
2.2 Vaccine design based upon the functions of human cytomegalovirus-encoded IL-10
3. Cytokine receptor homolog encoded by the human cytomegalovirus UL144 gene
4. Chemokine homologs encoded by human cytomegalovirus UL146, UL147 and UL128
4.1 UL146 and UL147
4.2. UL128
5. The chemokine homolog UL146 and the cytokine receptor homolog UL144 are hypervariable genes
6. Chemokine receptor family members encoded by human cytomegalovirus
6.1. US28
6.1.1. US28 immunomodulation
6.1.2. US28 in tumorigenesis
6.2. US27, UL33 and UL78
6.2.1. US27
6.2.2. UL33 and UL78
6.3. UL21.5
7. Conclusions
Acknowledgments
Conflict of Interest
References and Notes
- Slobedman, B.; Cao, J.Z.; Avdic, S.; Webster, B.; McAllery, S.; Cheung, A.K.; Tan, J.C.; Abendroth, A. Human cytomegalovirus latent infection and associated viral gene expression. Future Microbiol. 2010, 5, 883–900. [Google Scholar] [CrossRef]
- Mocarski, E.S.; Shenk, T.; Pass, R.F. Cytomegaloviruses, 5thKnipe, D.M., Howley, P.M., Eds.; Lippincott Williams & Wilkins: Philadelphia, PA, 2007; Volume 2, pp. 2701–2772. [Google Scholar]
- Sylwester, A.W.; Mitchell, B.L.; Edgar, J.B.; Taormina, C.; Pelte, C.; Ruchti, F.; Sleath, P.R.; Grabstein, K.H.; Hosken, N.A.; Kern, F.; Nelson, J.A.; Picker, L.J. Broadly targeted human cytomegalovirus-specific CD4+ and CD8+ T cells dominate the memory compartments of exposed subjects. J. Exp. Med. 2005, 202, 673–685. [Google Scholar] [CrossRef]
- Vescovini, R.; Biasini, C.; Fagnoni, F.F.; Telera, A.R.; Zanlari, L.; Pedrazzoni, M.; Bucci, L.; Monti, D.; Medici, M.C.; Chezzi, C.; Franceschi, C.; Sansoni, P. Massive load of functional effector CD4+ and CD8+ T cells against cytomegalovirus in very old subjects. J Immunol 2007, 179, 4283–4291. [Google Scholar]
- Kotenko, S.V.; Saccani, S.; Izotova, L.S.; Mirochnitchenko, O.V.; Pestka, S. Human cytomegalovirus harbors its own unique IL-10 homolog (cmvIL-10). Proc. Natl. Acad. Sci. U.S.A 2000, 97, 1695–1700. [Google Scholar] [CrossRef]
- Lockridge, K.M.; Zhou, S.S.; Kravitz, R.H.; Johnson, J.L.; Sawai, E.T.; Blewett, E.L.; Barry, P.A. Primate cytomegaloviruses encode and express an IL-10-like protein. Virology 2000, 268, 272–280. [Google Scholar] [CrossRef]
- Jenkins, C.; Abendroth, A.; Slobedman, B. A novel viral transcript with homology to human interleukin-10 is expressed during latent human cytomegalovirus infection. J. Virol. 2004, 78, 1440–1447. [Google Scholar] [CrossRef]
- Lin, Y.L.; Chang, P.C.; Wang, Y.; Li, M. Identification of novel viral interleukin-10 isoforms of human cytomegalovirus AD169. Virus Res. 2008, 131, 213–223. [Google Scholar] [CrossRef]
- Jones, B.C.; Logsdon, N.J.; Josephson, K.; Cook, J.; Barry, P.A.; Walter, M.R. Crystal structure of human cytomegalovirus IL-10 bound to soluble human IL-10R1. Proc. Natl. Acad. Sci. U.S.A. 2002, 99, 9404–9409. [Google Scholar]
- Nachtwey, J.; Spencer, J.V. HCMV IL-10 suppresses cytokine expression in monocytes through inhibition of nuclear factor-kappaB. Viral Immunol. 2008, 21, 477–482. [Google Scholar] [CrossRef]
- Spencer, J.V. The cytomegalovirus homolog of interleukin-10 requires phosphatidylinositol 3-kinase activity for inhibition of cytokine synthesis in monocytes. J. Virol. 2007, 81, 2083–2086. [Google Scholar] [CrossRef]
- Spencer, J.V.; Lockridge, K.M.; Barry, P.A.; Lin, G.; Tsang, M.; Penfold, M.E.; Schall, T.J. Potent immunosuppressive activities of cytomegalovirus-encoded interleukin-10. J. Virol. 2002, 76, 1285–1292. [Google Scholar]
- Chang, W.L.; Baumgarth, N.; Yu, D.; Barry, P.A. Human cytomegalovirus-encoded interleukin-10 homolog inhibits maturation of dendritic cells and alters their functionality. J. Virol. 2004, 78, 8720–8731. [Google Scholar] [CrossRef]
- Raftery, M.J.; Wieland, D.; Gronewald, S.; Kraus, A.A.; Giese, T.; Schonrich, G. Shaping phenotype, function, and survival of dendritic cells by cytomegalovirus-encoded IL-10. J Immunol 2004, 173, 3383–3391. [Google Scholar]
- Raftery, M.J.; Hitzler, M.; Winau, F.; Giese, T.; Plachter, B.; Kaufmann, S.H.; Schonrich, G. Inhibition of CD1 antigen presentation by human cytomegalovirus. J. Virol. 2008, 82, 4308–4319. [Google Scholar] [CrossRef]
- Chang, W.L.; Barry, P.A.; Szubin, R.; Wang, D.; Baumgarth, N. Human cytomegalovirus suppresses type I interferon secretion by plasmacytoid dendritic cells through its interleukin 10 homolog. Virology 2009, 390, 330–337. [Google Scholar] [CrossRef]
- Cheeran, M.C.; Hu, S.; Sheng, W.S.; Peterson, P.K.; Lokensgard, J.R. CXCL10 production from cytomegalovirus-stimulated microglia is regulated by both human and viral interleukin-10. J. Virol. 2003, 77, 4502–4515. [Google Scholar]
- Spencer, J.V.; Cadaoas, J.; Castillo, P.R.; Saini, V.; Slobedman, B. Stimulation of B lymphocytes by cmvIL-10 but not LAcmvIL-10. Virology 2008, 374, 164–169. [Google Scholar] [CrossRef]
- Jaworowski, A.; Cheng, W.J.; Westhorpe, C.L.; Abendroth, A.; Crowe, S.M.; Slobedman, B. Enhanced monocyte Fc phagocytosis by a homologue of interleukin-10 encoded by human cytomegalovirus. Virology 2009, 391, 20–24. [Google Scholar] [CrossRef]
- Yamamoto-Tabata, T.; McDonagh, S.; Chang, H.T.; Fisher, S.; Pereira, L. Human cytomegalovirus interleukin-10 downregulates metalloproteinase activity and impairs endothelial cell migration and placental cytotrophoblast invasiveness in vitro. J. Virol. 2004, 78, 2831–2840. [Google Scholar] [CrossRef]
- Jenkins, C.; Garcia, W.; Abendroth, A.; Slobedman, B. Expression of a human cytomegalovirus latency-associated homolog of interleukin-10 during the productive phase of infection. Virology 370, 285–294.
- Jenkins, C.; Garcia, W.; Godwin, M.J.; Spencer, J.V.; Stern, J.L.; Abendroth, A.; Slobedman, B. Immunomodulatory properties of a viral homolog of human interleukin-10 expressed by human cytomegalovirus during the latent phase of infection. J. Virol. 82, 3736–3750.
- Pepperl-Klindworth, S.; Besold, K.; Frankenberg, N.; Farkas, M.; Kuball, J.; Theobald, M.; Plachter, B. Cytomegalovirus interleukin-10 expression in infected cells does not impair MHC class I restricted Peptide presentation on bystanding antigen-presenting cells. Viral Immunol. 2006, 19, 92–101. [Google Scholar] [CrossRef]
- Cheung, A.K.; Gottlieb, D.J.; Plachter, B.; Pepperl-Klindworth, S.; Avdic, S.; Cunningham, A.L.; Abendroth, A.; Slobedman, B. The role of the human cytomegalovirus UL111A gene in downregulating CD4+ T cell recognition of latently infected cells: implications for virus elimination during latency. Blood 2009.
- Avdic, S.; Cao, J.Z.; Cheung, A.K.; Abendroth, A.; Slobedman, B. Viral interleukin-10 expressed by human cytomegalovirus during the latent phase of infection modulates latently infected myeloid cell differentiation. J. Virol. 2011, 85, 7465–7471. [Google Scholar] [CrossRef]
- Stratton, K.R.; Durch, J.S.; Lawrence, R.S. Vaccines for the 21st Century: A Tool for Decision making; The National Academies Press, 2000; p. 476. [Google Scholar]
- Khanna, R.; Diamond, D.J. Human cytomegalovirus vaccine: time to look for alternative options. Trends Mol. Med. 2006, 12, 26–33. [Google Scholar] [CrossRef]
- Schleiss, M.R. Cytomegalovirus vaccine development. Curr Top Microbiol Immunol 2008, 325, 361–382. [Google Scholar] [CrossRef]
- Logsdon, N.J.; Eberhardt, M.K.; Allen, C.E.; Barry, P.A.; Walter, M.R. Design and analysis of rhesus cytomegalovirus IL-10 mutants as a model for novel vaccines against human cytomegalovirus. PLoS One 2011, 6, e28127. [Google Scholar]
- Chang, W.L.; Barry, P.A. Attenuation of innate immunity by cytomegalovirus IL-10 establishes a long-term deficit of adaptive antiviral immunity. Proc. Natl. Acad. Sci. U.S.A. 2010, 107, 22647–22652. [Google Scholar]
- Yue, Y.; Barry, P.A. Rhesus cytomegalovirus a nonhuman primate model for the study of human cytomegalovirus. Adv. Virus Res. 2008, 72, 207–226. [Google Scholar] [CrossRef]
- Yue, Y.; Kaur, A.; Eberhardt, M.K.; Kassis, N.; Zhou, S.S.; Tarantal, A.F.; Barry, P.A. Immunogenicity and protective efficacy of DNA vaccines expressing rhesus cytomegalovirus glycoprotein B, phosphoprotein 65-2, and viral interleukin-10 in rhesus macaques. J. Virol. 2007, 81, 1095–1109. [Google Scholar]
- Eberhardt, M.K.; Chang, W.L.; Logsdon, N.J.; Yue, Y.; Walter, M.R.; Barry, P.A. Host immune responses to a viral immune modulating protein: immunogenicity of viral interleukin-10 in rhesus cytomegalovirus-infected rhesus macaques. PLoS One 2012, 7, e37931. [Google Scholar]
- de Lemos Rieper, C.; Galle, P.; Pedersen, B.K.; Hansen, M.B. Characterization of specific antibodies against cytomegalovirus (CMV)-encoded interleukin 10 produced by 28% of CMV-seropositive blood donors. J. Gen. Virol. 2011, 92, 1508–1518. [Google Scholar] [CrossRef]
- Walter, M.R.; Eberhardt, M.K.; Logsdon, N.J.; Allen, C.E.; Barry, P.A. PS2-111. Targeting the IL-10 signalling pathway as a vaccine strategy for HCMV. Cytokine 2011, 56, 94. [Google Scholar]
- Benedict, C.A.; Butrovich, K.D.; Lurain, N.S.; Corbeil, J.; Rooney, I.; Schneider, P.; Tschopp, J.; Ware, C.F. Cutting edge: a novel viral TNF receptor superfamily member in virulent strains of human cytomegalovirus. J Immunol 1999, 162, 6967–6970. [Google Scholar]
- Poole, E.; King, C.A.; Sinclair, J.H.; Alcami, A. The UL144 gene product of human cytomegalovirus activates NFkappaB via a TRAF6-dependent mechanism. EMBO J 2006, 25, 4390–4399. [Google Scholar] [CrossRef]
- Cheung, T.C.; Humphreys, I.R.; Potter, K.G.; Norris, P.S.; Shumway, H.M.; Tran, B.R.; Patterson, G.; Jean-Jacques, R.; Yoon, M.; Spear, P.G.; Murphy, K.M.; Lurain, N.S.; Benedict, C.A.; Ware, C.F. Evolutionarily divergent herpesviruses modulate T cell activation by targeting the herpesvirus entry mediator cosignaling pathway. Proc. Natl. Acad. Sci. U.S.A. 2005, 102, 13218–13223. [Google Scholar]
- Murphy, K.M.; Nelson, C.A.; Sedy, J.R. Balancing co-stimulation and inhibition with BTLA and HVEM. Nat Rev Immunol 2006, 6, 671–681. [Google Scholar] [CrossRef]
- Poole, E.; Groves, I.; MacDonald, A.; Pang, Y.; Alcami, A.; Sinclair, J. Identification of TRIM23 as a cofactor involved in the regulation of NF-kappaB by human cytomegalovirus. J. Virol. 2009, 83, 3581–3590. [Google Scholar] [CrossRef]
- Poole, E.; Atkins, E.; Nakayama, T.; Yoshie, O.; Groves, I.; Alcami, A.; Sinclair, J. NF-kappaB-mediated activation of the chemokine CCL22 by the product of the human cytomegalovirus gene UL144 escapes regulation by viral IE86. J. Virol. 2008, 82, 4250–4256. [Google Scholar] [CrossRef]
- Iellem, A.; Mariani, M.; Lang, R.; Recalde, H.; Panina-Bordignon, P.; Sinigaglia, F.; D'Ambrosio, D. Unique chemotactic response profile and specific expression of chemokine receptors CCR4 and CCR8 by CD4(+)CD25(+) regulatory T cells. J Exp Med 2001, 194, 847–853. [Google Scholar] [CrossRef]
- Zlotnik, A.; Yoshie, O. Chemokines: a new classification system and their role in immunity. Immunity 2000, 12, 121–127. [Google Scholar] [CrossRef]
- Penfold, M.E.; Dairaghi, D.J.; Duke, G.M.; Saederup, N.; Mocarski, E.S.; Kemble, G.W.; Schall, T.J. Cytomegalovirus encodes a potent alpha chemokine. Proc. Natl. Acad. Sci. U.S.A. 1999, 96, 9839–9844. [Google Scholar]
- Kobayashi, Y. The role of chemokines in neutrophil biology. Front Biosci 2008, 13, 2400–2407. [Google Scholar] [CrossRef]
- Luttichau, H.R. The cytomegalovirus UL146 gene product vCXCL1 targets both CXCR1 and CXCR2 as an agonist. J Biol Chem 2010, 285, 9137–9146. [Google Scholar] [CrossRef]
- Revello, M.G.; Gerna, G. Human cytomegalovirus tropism for endothelial/epithelial cells: scientific background and clinical implications. Rev Med Virol 2010, 20, 136–155. [Google Scholar] [CrossRef]
- Dargan, D.J.; Douglas, E.; Cunningham, C.; Jamieson, F.; Stanton, R.J.; Baluchova, K.; McSharry, B.P.; Tomasec, P.; Emery, V.C.; Percivalle, E.; Sarasini, A.; Gerna, G.; Wilkinson, G.W.; Davison, A.J. Sequential mutations associated with adaptation of human cytomegalovirus to growth in cell culture. J. Gen. Virol. 2010, 91, 1535–1546. [Google Scholar]
- Akter, P.; Cunningham, C.; McSharry, B.P.; Dolan, A.; Addison, C.; Dargan, D.J.; Hassan-Walker, A.F.; Emery, V.C.; Griffiths, P.D.; Wilkinson, G.W.; Davison, A.J. Two novel spliced genes in human cytomegalovirus. J. Gen. Virol. 2003, 84, 1117–1122. [Google Scholar]
- Zheng, Q.; Tao, R.; Gao, H.; Xu, J.; Shang, S.; Zhao, N. HCMV-encoded UL128 enhances TNF-alpha and IL-6 expression and promotes PBMC proliferation through the MAPK/ERK pathway in vitro. Viral Immunol. 2012, 25, 98–105. [Google Scholar] [CrossRef]
- Fischer, F.R.; Luo, Y.; Luo, M.; Santambrogio, L.; Dorf, M.E. RANTES-induced chemokine cascade in dendritic cells. J. Immunol. 2001, 167, 1637–1643. [Google Scholar]
- Cha, T.A.; Tom, E.; Kemble, G.W.; Duke, G.M.; Mocarski, E.S.; Spaete, R.R. Human cytomegalovirus clinical isolates carry at least 19 genes not found in laboratory strains. J. Virol. 1996, 70, 78–83. [Google Scholar]
- Pignatelli, S.; Dal Monte, P.; Rossini, G.; Landini, M.P. Genetic polymorphisms among human cytomegalovirus (HCMV) wild-type strains. Rev. Med. Virol. 2004, 14, 383–410. [Google Scholar] [CrossRef]
- Dolan, A.; Cunningham, C.; Hector, R.D.; Hassan-Walker, A.F.; Lee, L.; Addison, C.; Dargan, D.J.; McGeoch, D.J.; Gatherer, D.; Emery, V.C.; Griffiths, P.D.; Sinzger, C.; McSharry, B.P.; Wilkinson, G.W.; Davison, A.J. Genetic content of wild-type human cytomegalovirus. J. Gen. Virol. 2004, 85, 1301–1312. [Google Scholar] [CrossRef]
- Heo, J.; Petheram, S.; Demmler, G.; Murph, J.R.; Adler, S.P.; Bale, J.; Sparer, T.E. Polymorphisms within human cytomegalovirus chemokine (UL146/UL147) and cytokine receptor genes (UL144) are not predictive of sequelae in congenitally infected children. Virology 2008, 378, 86–96. [Google Scholar] [CrossRef]
- Mao, Z.Q.; He, R.; Sun, M.; Qi, Y.; Huang, Y.J.; Ruan, Q. The relationship between polymorphisms of HCMV UL144 ORF and clinical manifestations in 73 strains with congenital and/or perinatal HCMV infection. Arch. Virol. 2007, 152, 115–124. [Google Scholar] [CrossRef]
- Picone, O.; Costa, J.M.; Chaix, M.L.; Ville, Y.; Rouzioux, C.; Leruez-Ville, M. Human cytomegalovirus UL144 gene polymorphisms in congenital infections. J. Clin. Microbiol. 2005, 43, 25–29. [Google Scholar] [CrossRef]
- Yan, H.; Koyano, S.; Inami, Y.; Yamamoto, Y.; Suzutani, T.; Mizuguchi, M.; Ushijima, H.; Kurane, I.; Inoue, N. Genetic variations in the gB, UL144 and UL149 genes of human cytomegalovirus strains collected from congenitally and postnatally infected Japanese children. Arch. Virol. 2008, 153, 667–674. [Google Scholar] [CrossRef]
- Paradowska, E.; Studzinska, M.; Nowakowska, D.; Wilczynski, J.; Rycel, M.; Suski, P.; Gaj, Z.; Kaczmarek, B.; Zbrog, Z.; Lesnikowski, Z.J. Distribution of UL144, US28 and UL55 genotypes in Polish newborns with congenital cytomegalovirus infections. Eur J. Clin. Microbiol. Infect Dis 2012, 31, 1335–1345. [Google Scholar] [CrossRef]
- Waters, A.; Hassan, J.; De Gascun, C.; Kissoon, G.; Knowles, S.; Molloy, E.; Connell, J.; Hall, W.W. Human cytomegalovirus UL144 is associated with viremia and infant development sequelae in congenital infection. J. Clin. Microbiol. 2010, 48, 3956–3962. [Google Scholar] [CrossRef]
- Arav-Boger, R.; Willoughby, R.E.; Pass, R.F.; Zong, J.C.; Jang, W.J.; Alcendor, D.; Hayward, G.S. Polymorphisms of the cytomegalovirus (CMV)-encoded tumor necrosis factor-alpha and beta-chemokine receptors in congenital CMV disease. J. Infect. Dis. 2002, 186, 1057–1064. [Google Scholar] [CrossRef]
- Arav-Boger, R.; Foster, C.B.; Zong, J.C.; Pass, R.F. Human cytomegalovirus-encoded alpha -chemokines exhibit high sequence variability in congenitally infected newborns. J. Infect. Dis. 2006, 193, 788–791. [Google Scholar] [CrossRef]
- Prichard, M.N.; Penfold, M.E.; Duke, G.M.; Spaete, R.R.; Kemble, G.W. A review of genetic differences between limited and extensively passaged human cytomegalovirus strains. Rev. Med. Virol. 2001, 11, 191–200. [Google Scholar] [CrossRef]
- Arav-Boger, R.; Boger, Y.S.; Foster, C.B.; Boger, Z. The use of artificial neural networks in prediction of congenital CMV outcome from sequence data. Bioinform. Biol. Insights. 2008, 2, 281–289. [Google Scholar]
- Rosenbaum, D.M.; Rasmussen, S.G.; Kobilka, B.K. The structure and function of G-protein-coupled receptors. Nature 2009, 459, 356–363. [Google Scholar] [CrossRef]
- Allen, S.J.; Crown, S.E.; Handel, T.M. Chemokine: receptor structure, interactions, and antagonism. Annu. Rev. Immunol. 2007, 25, 787–820. [Google Scholar] [CrossRef]
- Chee, M.S.; Satchwell, S.C.; Preddie, E.; Weston, K.M.; Barrell, B.G. Human cytomegalovirus encodes three G protein-coupled receptor homologues. Nature 1990, 344, 774–777. [Google Scholar] [CrossRef]
- Gompels, U.A.; Nicholas, J.; Lawrence, G.; Jones, M.; Thomson, B.J.; Martin, M.E.; Efstathiou, S.; Craxton, M.; Macaulay, H.A. The DNA sequence of human herpesvirus-6: structure, coding content, and genome evolution. Virology 1995, 209, 29–51. [Google Scholar] [CrossRef]
- Vomaske, J.; Nelson, J.A.; Streblow, D.N. Human Cytomegalovirus US28: a functionally selective chemokine binding receptor. Infect. Disord. Drug Targets 2009, 9, 548–556. [Google Scholar] [CrossRef]
- Boomker, J.M.; van Luyn, M.J.; The, T.H.; de Leij, L.F.; Harmsen, M.C. US28 actions in HCMV infection: lessons from a versatile hijacker. Rev. Med. Virol. 2005, 15, 269–282. [Google Scholar] [CrossRef]
- Vischer, H.F.; Leurs, R.; Smit, M.J. HCMV-encoded G-protein-coupled receptors as constitutively active modulators of cellular signaling networks. Trends Pharmacol. Sci. 2006, 27, 56–63. [Google Scholar] [CrossRef]
- Zipeto, D.; Bodaghi, B.; Laurent, L.; Virelizier, J.L.; Michelson, S. Kinetics of transcription of human cytomegalovirus chemokine receptor US28 in different cell types. J. Gen. Virol. 1999, 80 ( Pt 3), 543–547. [Google Scholar]
- Gao, J.L.; Murphy, P.M. Human cytomegalovirus open reading frame US28 encodes a functional beta chemokine receptor. J. Biol. Chem. 1994, 269, 28539–28542. [Google Scholar]
- Neote, K.; DiGregorio, D.; Mak, J.Y.; Horuk, R.; Schall, T.J. Molecular cloning, functional expression, and signaling characteristics of a C-C chemokine receptor. Cell 1993, 72, 415–425. [Google Scholar] [CrossRef]
- Kledal, T.N.; Rosenkilde, M.M.; Schwartz, T.W. Selective recognition of the membrane-bound CX3C chemokine, fractalkine, by the human cytomegalovirus-encoded broad-spectrum receptor US28. FEBS Lett 1998, 441, 209–214. [Google Scholar] [CrossRef]
- Streblow, D.N.; Vomaske, J.; Smith, P.; Melnychuk, R.; Hall, L.; Pancheva, D.; Smit, M.; Casarosa, P.; Schlaepfer, D.D.; Nelson, J.A. Human cytomegalovirus chemokine receptor US28-induced smooth muscle cell migration is mediated by focal adhesion kinase and Src. J. Biol. Chem. 2003, 278, 50456–50465. [Google Scholar]
- Billstrom, M.A.; Johnson, G.L.; Avdi, N.J.; Worthen, G.S. Intracellular signaling by the chemokine receptor US28 during human cytomegalovirus infection. J. Virol. 1998, 72, 5535–5544. [Google Scholar]
- Vieira, J.; Schall, T.J.; Corey, L.; Geballe, A.P. Functional analysis of the human cytomegalovirus US28 gene by insertion mutagenesis with the green fluorescent protein gene. J. Virol. 1998, 72, 8158–8165. [Google Scholar]
- Stropes, M.P.; Schneider, O.D.; Zagorski, W.A.; Miller, J.L.; Miller, W.E. The carboxy-terminal tail of human cytomegalovirus (HCMV) US28 regulates both chemokine-independent and chemokine-dependent signaling in HCMV-infected cells. J. Virol. 2009, 83, 10016–10027. [Google Scholar] [CrossRef]
- Minisini, R.; Tulone, C.; Luske, A.; Michel, D.; Mertens, T.; Gierschik, P.; Moepps, B. Constitutive inositol phosphate formation in cytomegalovirus-infected human fibroblasts is due to expression of the chemokine receptor homologue pUS28. J. Virol. 2003, 77, 4489–4501. [Google Scholar] [CrossRef]
- Casarosa, P.; Gruijthuijsen, Y.K.; Michel, D.; Beisser, P.S.; Holl, J.; Fitzsimons, C.P.; Verzijl, D.; Bruggeman, C.A.; Mertens, T.; Leurs, R.; Vink, C.; Smit, M.J. Constitutive signaling of the human cytomegalovirus-encoded receptor UL33 differs from that of its rat cytomegalovirus homolog R33 by promiscuous activation of G proteins of the Gq, Gi, and Gs classes. J. Biol. Chem. 2003, 278, 50010–50023. [Google Scholar]
- Waldhoer, M.; Kledal, T.N.; Farrell, H.; Schwartz, T.W. Murine cytomegalovirus (CMV) M33 and human CMV US28 receptors exhibit similar constitutive signaling activities. J. Virol. 2002, 76, 8161–8168. [Google Scholar] [CrossRef]
- Casarosa, P.; Bakker, R.A.; Verzijl, D.; Navis, M.; Timmerman, H.; Leurs, R.; Smit, M.J. Constitutive signaling of the human cytomegalovirus-encoded chemokine receptor US28. J. Biol. Chem. 2001, 276, 1133–1137. [Google Scholar]
- Miller, W.E.; Houtz, D.A.; Nelson, C.D.; Kolattukudy, P.E.; Lefkowitz, R.J. G-protein-coupled receptor (GPCR) kinase phosphorylation and beta-arrestin recruitment regulate the constitutive signaling activity of the human cytomegalovirus US28 GPCR. J. Biol. Chem. 2003, 278, 21663–21671. [Google Scholar]
- Caposio, P.; Orloff, S.L.; Streblow, D.N. The role of cytomegalovirus in angiogenesis. Virus Res. 2008, 157, 204–211. [Google Scholar]
- Farrell, H.E.; Abraham, A.M.; Cardin, R.D.; Sparre-Ulrich, A.H.; Rosenkilde, M.M.; Spiess, K.; Jensen, T.H.; Kledal, T.N.; Davis-Poynter, N. Partial functional complementation between human and mouse cytomegalovirus chemokine receptor homologues. J. Virol. 2011, 85, 6091–6095. [Google Scholar]
- Davis-Poynter, N.J.; Lynch, D.M.; Vally, H.; Shellam, G.R.; Rawlinson, W.D.; Barrell, B.G.; Farrell, H.E. Identification and characterization of a G protein-coupled receptor homolog encoded by murine cytomegalovirus. J. Virol. 1997, 71, 1521–1529. [Google Scholar]
- Hargett, D.; Shenk, T.E. Experimental human cytomegalovirus latency in CD14+ monocytes. Proc. Natl. Acad. Sci. U.S.A. 2010, 107, 20039–20044. [Google Scholar] [CrossRef]
- Beisser, P.S.; Laurent, L.; Virelizier, J.L.; Michelson, S. Human cytomegalovirus chemokine receptor gene US28 is transcribed in latently infected THP-1 monocytes. J. Virol. 2001, 75, 5949–5957. [Google Scholar] [CrossRef]
- Cheung, A.K.; Abendroth, A.; Cunningham, A.L.; Slobedman, B. Viral gene expression during the establishment of human cytomegalovirus latent infection in myeloid progenitor cells. Blood 2006, 108, 3691–3699. [Google Scholar] [CrossRef]
- Bodaghi, B.; Jones, T.R.; Zipeto, D.; Vita, C.; Sun, L.; Laurent, L.; Arenzana-Seisdedos, F.; Virelizier, J.L.; Michelson, S. Chemokine sequestration by viral chemoreceptors as a novel viral escape strategy: withdrawal of chemokines from the environment of cytomegalovirus-infected cells. J. Exp. Med. 1998, 188, 855–866. [Google Scholar] [CrossRef]
- Billstrom, M.A.; Lehman, L.A.; Scott Worthen, G. Depletion of extracellular RANTES during human cytomegalovirus infection of endothelial cells. Am J Respir Cell Mol Biol 1999, 21, 163–167. [Google Scholar]
- Randolph-Habecker, J.R.; Rahill, B.; Torok-Storb, B.; Vieira, J.; Kolattukudy, P.E.; Rovin, B.H.; Sedmak, D.D. The expression of the cytomegalovirus chemokine receptor homolog US28 sequesters biologically active CC chemokines and alters IL-8 production. Cytokine 2002, 19, 37–46. [Google Scholar] [CrossRef]
- Boomker, J.M.; de Jong, E.K.; de Leij, L.F.; Harmsen, M.C. Chemokine scavenging by the human cytomegalovirus chemokine decoy receptor US28 does not inhibit monocyte adherence to activated endothelium. Antiviral Res. 2006, 69, 124–127. [Google Scholar] [CrossRef]
- Soroceanu, L.; Cobbs, C.S. Is HCMV a tumor promoter? Virus Res. 2011, 157, 193–203. [Google Scholar] [CrossRef]
- Maussang, D.; Verzijl, D.; van Walsum, M.; Leurs, R.; Holl, J.; Pleskoff, O.; Michel, D.; van Dongen, G.A.; Smit, M.J. Human cytomegalovirus-encoded chemokine receptor US28 promotes tumorigenesis. Proc. Natl. Acad. Sci. U.S.A. 2006, 103, 13068–13073. [Google Scholar]
- Maussang, D.; Langemeijer, E.; Fitzsimons, C.P.; Stigter-van Walsum, M.; Dijkman, R.; Borg, M.K.; Slinger, E.; Schreiber, A.; Michel, D.; Tensen, C.P.; van Dongen, G.A.; Leurs, R.; Smit, M.J. The human cytomegalovirus-encoded chemokine receptor US28 promotes angiogenesis and tumor formation via cyclooxygenase-2. Cancer Res. 2009, 69, 2861–2869. [Google Scholar]
- Slinger, E.; Maussang, D.; Schreiber, A.; Siderius, M.; Rahbar, A.; Fraile-Ramos, A.; Lira, S.A.; Soderberg-Naucler, C.; Smit, M.J. HCMV-encoded chemokine receptor US28 mediates proliferative signaling through the IL-6-STAT3 axis. Sci. Signal. 2010, 3, ra58. [Google Scholar] [CrossRef]
- Bongers, G.; Maussang, D.; Muniz, L.R.; Noriega, V.M.; Fraile-Ramos, A.; Barker, N.; Marchesi, F.; Thirunarayanan, N.; Vischer, H.F.; Qin, L.; Mayer, L.; Harpaz, N.; Leurs, R.; Furtado, G.C.; Clevers, H.; Tortorella, D.; Smit, M.J.; Lira, S.A. The cytomegalovirus-encoded chemokine receptor US28 promotes intestinal neoplasia in transgenic mice. J. Clin. Invest. 2010, 120, 3969–3978. [Google Scholar]
- Soroceanu, L.; Matlaf, L.; Bezrookove, V.; Harkins, L.; Martinez, R.; Greene, M.; Soteropoulos, P.; Cobbs, C.S. Human cytomegalovirus US28 found in glioblastoma promotes an invasive and angiogenic phenotype. Cancer Res. 2011, 71, 6643–6653. [Google Scholar] [CrossRef]
- Margulies, B.J.; Gibson, W. The chemokine receptor homologue encoded by US27 of human cytomegalovirus is heavily glycosylated and is present in infected human foreskin fibroblasts and enveloped virus particles. Virus Res. 2007, 123, 57–71. [Google Scholar] [CrossRef]
- O'Connor, C.M.; Shenk, T. Human cytomegalovirus pUS27 G protein-coupled receptor homologue is required for efficient spread by the extracellular route but not for direct cell-to-cell spread. J. Virol. 2011, 85, 3700–3707. [Google Scholar] [CrossRef]
- Fraile-Ramos, A.; Pelchen-Matthews, A.; Kledal, T.N.; Browne, H.; Schwartz, T.W.; Marsh, M. Localization of HCMV UL33 and US27 in endocytic compartments and viral membranes. Traffic 2002, 3, 218–232. [Google Scholar] [CrossRef]
- Tschische, P.; Tadagaki, K.; Kamal, M.; Jockers, R.; Waldhoer, M. Heteromerization of human cytomegalovirus encoded chemokine receptors. Biochem. Pharmacol. 2011, 82, 610–619. [Google Scholar] [CrossRef] [Green Version]
- Michel, D.; Milotic, I.; Wagner, M.; Vaida, B.; Holl, J.; Ansorge, R.; Mertens, T. The human cytomegalovirus UL78 gene is highly conserved among clinical isolates, but is dispensable for replication in fibroblasts and a renal artery organ-culture system. J. Gen. Virol. 2005, 86, 297–306. [Google Scholar] [CrossRef]
- Margulies, B.J.; Browne, H.; Gibson, W. Identification of the human cytomegalovirus G protein-coupled receptor homologue encoded by UL33 in infected cells and enveloped virus particles. Virology 1996, 225, 111–125. [Google Scholar] [CrossRef]
- Wagner, S.; Arnold, F.; Wu, Z.; Schubert, A.; Walliser, C.; Tadagaki, K.; Jockers, R.; Mertens, T.; Michel, D. The 7-transmembrane protein homologue UL78 of the human cytomegalovirus forms oligomers and traffics between the plasma membrane and different intracellular compartments. Arch. Virol. 2012, 157, 935–949. [Google Scholar] [CrossRef]
- Tadagaki, K.; Tudor, D.; Gbahou, F.; Tschische, P.; Waldhoer, M.; Bomsel, M.; Jockers, R.; Kamal, M. Human cytomegalovirus-encoded UL33 and UL78 heteromerize with host CCR5 and CXCR4 impairing their HIV coreceptor activity. Blood 2012, 119, 4908–4918. [Google Scholar] [CrossRef]
- Wang, D.; Bresnahan, W.; Shenk, T. Human cytomegalovirus encodes a highly specific RANTES decoy receptor. Proc. Natl. Acad. Sci. U.S.A. 2004, 101, 16642–16647. [Google Scholar] [CrossRef]
- Bresnahan, W.A.; Shenk, T. A subset of viral transcripts packaged within human cytomegalovirus particles. Science 2000, 288, 2373–2376. [Google Scholar] [CrossRef]
- Mortier, A.; Van Damme, J.; Proost, P. Regulation of chemokine activity by posttranslational modification. Pharmacol. Ther. 2008, 120, 197–217. [Google Scholar] [CrossRef]
- Taylor, R.T.; Bresnahan, W.A. Human cytomegalovirus immediate-early 2 protein IE86 blocks virus-induced chemokine expression. J. Virol. 2006, 80, 920–928. [Google Scholar] [CrossRef]
- Kim, Y.; Lee, S.; Kim, S.; Kim, D.; Ahn, J.H.; Ahn, K. Human cytomegalovirus clinical strain-specific microRNA miR-UL148D targets the human chemokine RANTES during infection. PLoS Pathog. 2012, 8, e1002577. [Google Scholar] [CrossRef]
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
McSharry, B.P.; Avdic, S.; Slobedman, B. Human Cytomegalovirus Encoded Homologs of Cytokines, Chemokines and their Receptors: Roles in Immunomodulation. Viruses 2012, 4, 2448-2470. https://doi.org/10.3390/v4112448
McSharry BP, Avdic S, Slobedman B. Human Cytomegalovirus Encoded Homologs of Cytokines, Chemokines and their Receptors: Roles in Immunomodulation. Viruses. 2012; 4(11):2448-2470. https://doi.org/10.3390/v4112448
Chicago/Turabian StyleMcSharry, Brian P., Selmir Avdic, and Barry Slobedman. 2012. "Human Cytomegalovirus Encoded Homologs of Cytokines, Chemokines and their Receptors: Roles in Immunomodulation" Viruses 4, no. 11: 2448-2470. https://doi.org/10.3390/v4112448
APA StyleMcSharry, B. P., Avdic, S., & Slobedman, B. (2012). Human Cytomegalovirus Encoded Homologs of Cytokines, Chemokines and their Receptors: Roles in Immunomodulation. Viruses, 4(11), 2448-2470. https://doi.org/10.3390/v4112448





