Adenovirus with DNA Packaging Gene Mutations Increased Virus Release
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Lines and Culture Conditions
2.2. Adenoviral Vectors
2.3. Virus Titration and Release
2.4. Cytotoxicity Assay
2.5. DNA Sequencing
2.6. Lung Cancer Xenograft
2.7. Immunohistochemistry
2.8. Statistical Analysis
3. Results
3.1. AdUV with Wildtype E1 and Mutated E3
3.2. Mutations That Affect Ad Packaging
3.3. Mutations Affecting Other Ad Structural Proteins
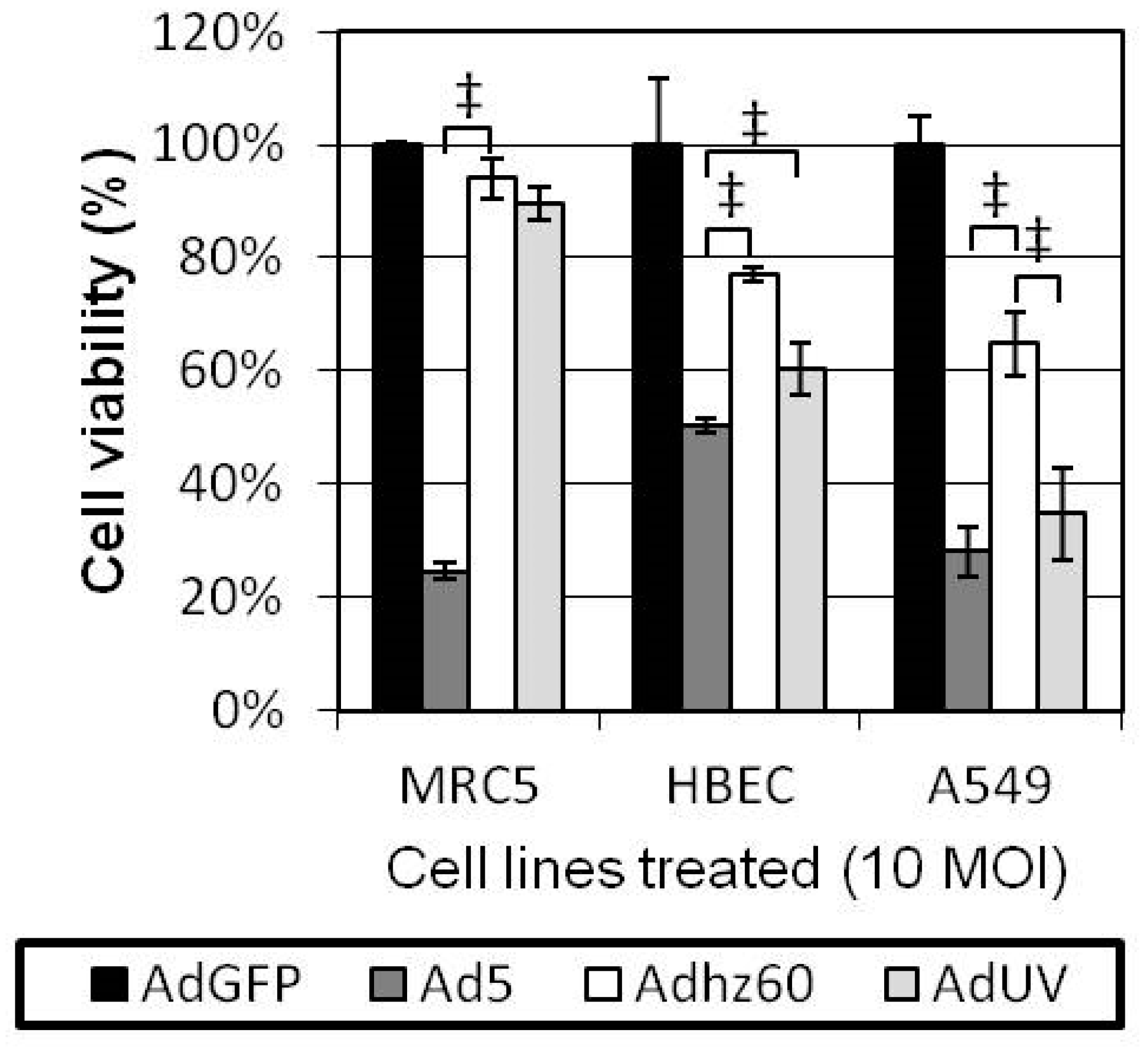
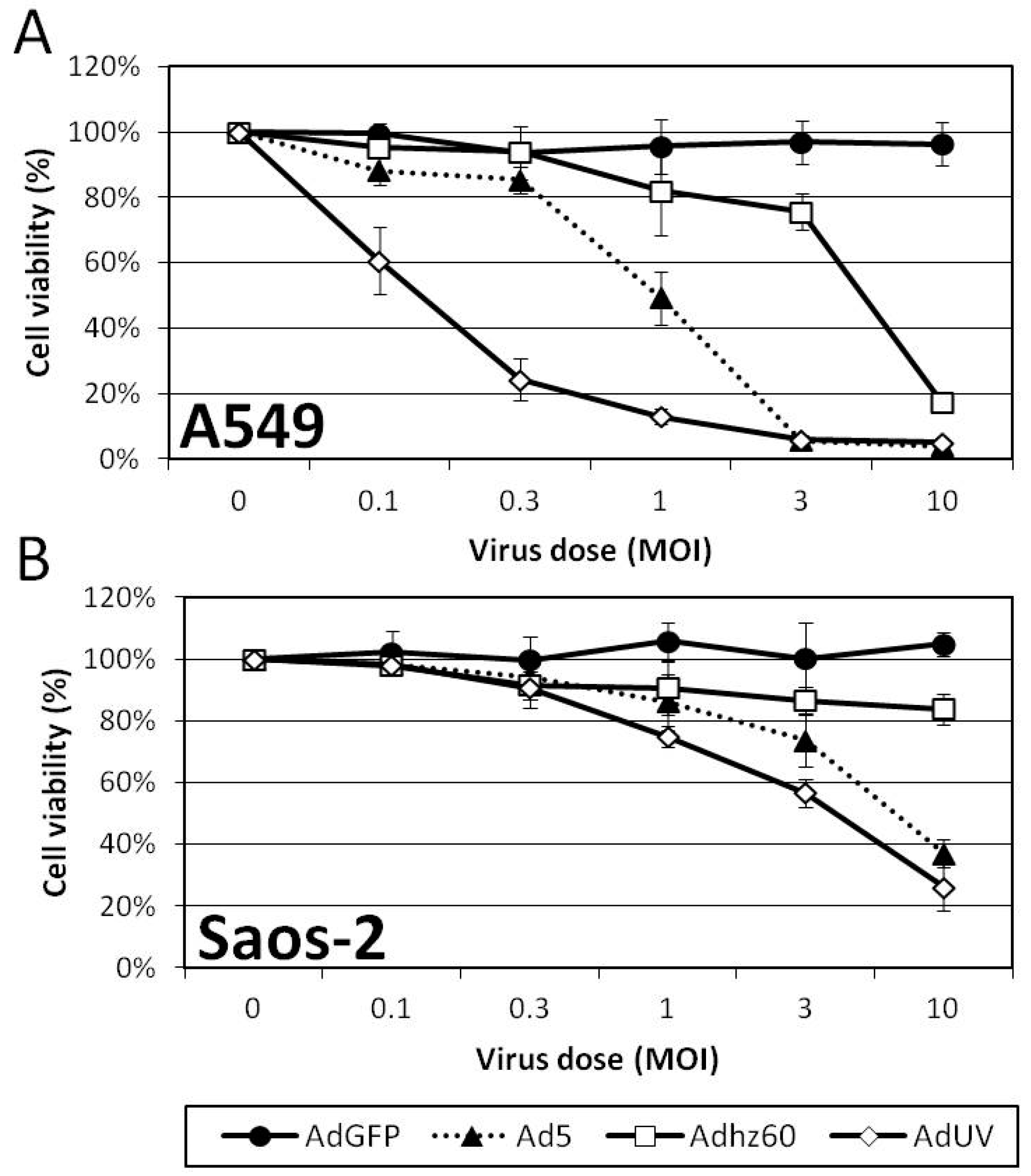
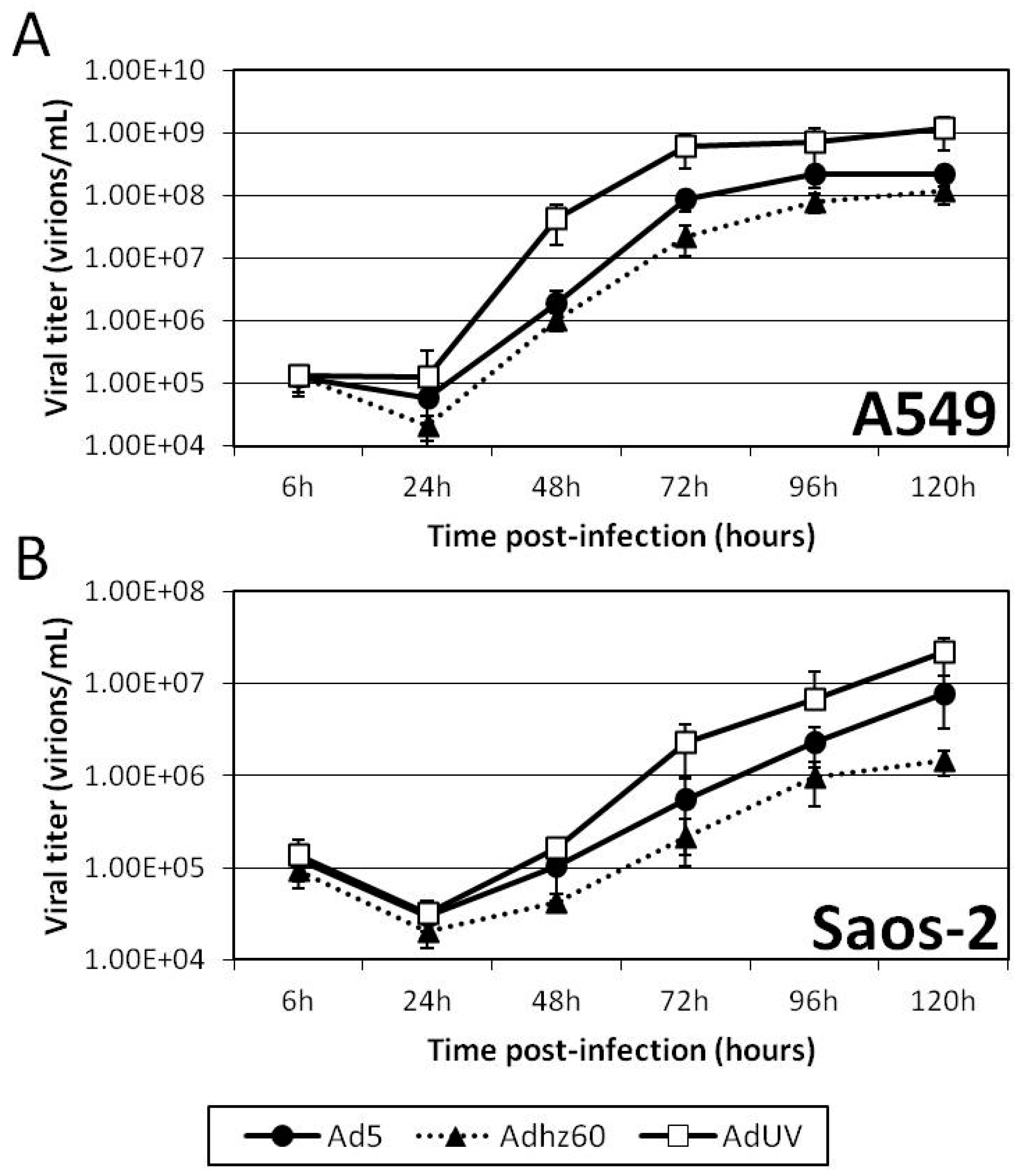
3.4. AdUV Displayed Greater Oncolysis and Release from A549 and Saos-2 Cells
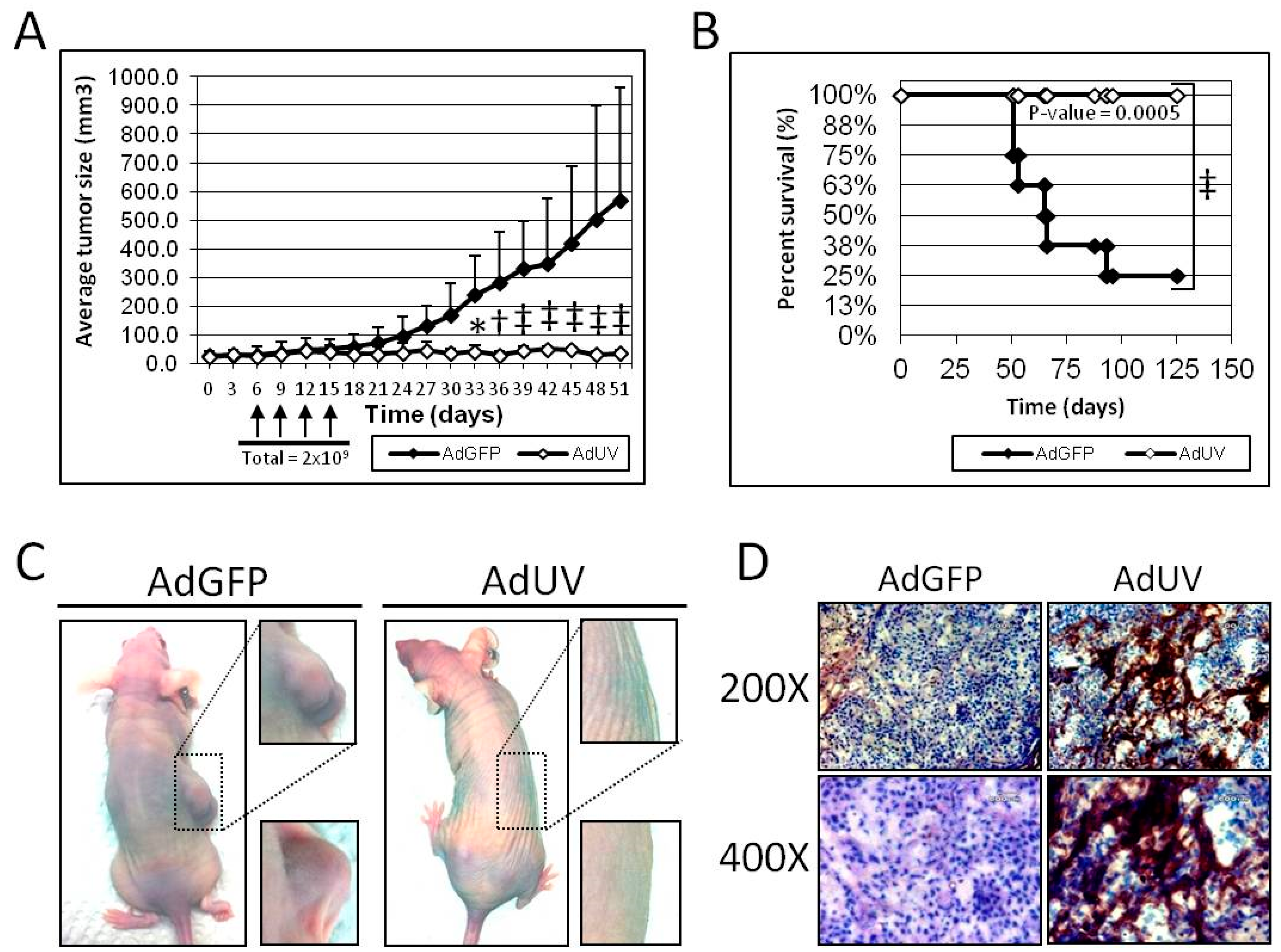
3.5. AdUV Inhibits Tumor Growth in Nude Mice
4. Discussion
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Mulvihill, S.; Warren, R.; Venook, A.; Adler, A.; Randlev, B.; Heise, C.; Kirn, D. Safety and feasibility of injection with an E1B-55 kDa gene-deleted, replication-selective adenovirus (ONYX-015) into primary carcinomas of the pancreas: a phase I trial. Gene Ther. 2001, 8, 308–315. [Google Scholar] [CrossRef] [PubMed]
- Ko, D.; Hawkins, L.; Yu, D.C. Development of transcriptionally regulated oncolytic adenoviruses. Oncogene 2005, 24, 7763–7774. [Google Scholar] [CrossRef] [PubMed]
- Berk, A.J. Adenoviridae: The Viruses and Their Replication. In Fields Virology; Knipe, D.M., Howley, P.M., Eds.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2007; pp. 2355–2394. [Google Scholar]
- Lu, W.; Zheng, S.; Li, X.F.; Huang, J.J.; Zheng, X.; Li, Z. Intra-tumor injection of H101, a recombinant adenovirus, in combination with chemotherapy in patients with advanced cancers: A pilot phase II clinical trial. World J. Gastroenterol. 2004, 10, 3634–3638. [Google Scholar] [CrossRef]
- Ye, W.; Liu, R.; Pan, C.; Jiang, W.; Zhang, L.; Guan, Z.; Wu, J.; Ying, X.; Li, L.; Li, S.; et al. Multicenter randomized phase 2 clinical trial of a recombinant human endostatin adenovirus in patients with advanced head and neck carcinoma. Mol. Ther. 2014, 22, 1221–1229. [Google Scholar] [CrossRef] [PubMed]
- Parker, J.N.; Bauer, D.F.; Cody, J.J.; Markert, J.M. Oncolytic viral therapy of malignant glioma. Neurotherapeutics 2009, 6, 558–569. [Google Scholar] [CrossRef] [PubMed]
- Nettelbeck, D.M. Cellular genetic tools to control oncolytic adenoviruses for virotherapy of cancer. J. Mol. Med. 2008, 86, 363–377. [Google Scholar] [CrossRef] [PubMed]
- Zhao, T.; Rao, X.M.; Xie, X.; Li, L.; Thompson, T.C.; McMasters, K.M.; Zhou, H.S. Adenovirus with insertion-mutated E1A selectively propagates in liver cancer cells and destroys tumors in vivo. Cancer Res. 2003, 63, 3073–3078. [Google Scholar] [PubMed]
- Cheng, P.H.; Rao, X.M.; Wechman, S.L.; Li, X.F.; McMasters, K.M.; Zhou, H.S. Oncolytic adenovirus targeting cyclin E overexpression repressed tumor growth in syngeneic immunocompetent mice. BMC Cancer 2015, 15, 716. [Google Scholar] [CrossRef] [PubMed]
- Parato, K.A.; Senger, D.; Forsyth, P.A.; Bell, J.C. Recent progress in the battle between oncolytic viruses and tumours. Nat. Rev. Cancer 2005, 5, 965–976. [Google Scholar] [CrossRef] [PubMed]
- Smith, E.; Breznik, J.; Lichty, B.D. Strategies to enhance viral penetration of solid tumors. Hum. Gene Ther. 2011, 22, 1053–1060. [Google Scholar] [CrossRef] [PubMed]
- Galanis, E.; Okuno, S.H.; Nascimento, A.G.; Lewis, B.D.; Lee, R.A.; Oliveira, A.M.; Sloan, J.A.; Atherton, P.; Edmonson, J.H.; Erlichman, C.; et al. Phase I-II trial of ONYX-015 in combination with MAP chemotherapy in patients with advanced sarcomas. Gene Ther. 2005, 12, 437–445. [Google Scholar] [CrossRef] [PubMed]
- Seth, P.; Higginbotham, J. Advantages and disadvantages of multiple different methods of adenoviral vector construction. Methods Mol. Med. 2000, 45, 189–198. [Google Scholar] [PubMed]
- Ben-Israel, H.; Kleinberger, T. Adenovirus and cell cycle control. Front. Biosci. 2002, 7, d1369–d1395. [Google Scholar] [CrossRef]
- Barker, D.D.; Berk, A.J. Adenovirus proteins from both E1B reading frames are required for transformation of rodent cells by viral infection and DNA transfection. Virology 1987, 156, 107–121. [Google Scholar] [CrossRef]
- Stillman, B. Functions of the adenovirus E1B tumour antigens. Cancer Surv. 1986, 5, 389–404. [Google Scholar] [PubMed]
- Lowe, S.W.; Ruley, H.E. Stabilization of the p53 tumor suppressor is induced by adenovirus 5 E1A and accompanies apoptosis. Genes Dev. 1993, 7, 535–545. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Rao, X.M.; Snodgrass, C.L.; McMasters, K.M.; Zhou, H.S. Selective replication of E1B55K-deleted adenoviruses depends on enhanced E1A expression in cancer cells. Cancer Gene Ther. 2006, 13, 572–583. [Google Scholar] [CrossRef] [PubMed]
- Rao, X.M.; Zheng, X.; Waigel, S.; Zacharias, W.; McMasters, K.M.; Zhou, H.S. Gene expression profiles of normal human lung cells affected by adenoviral E1B. Virology 2006, 350, 418–428. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Rao, X.M.; Gomez-Gutierrez, J.G.; Hao, H.; McMasters, K.M.; Zhou, H.S. Adenovirus E1B55K region is required to enhance cyclin E expression for efficient viral DNA replication. J. Virol. 2008, 82, 3415–3427. [Google Scholar] [CrossRef] [PubMed]
- Cheng, P.H.; Wechman, S.L.; McMasters, K.M.; Zhou, H.S. Oncolytic Replication of E1b-Deleted Adenoviruses. Viruses 2015, 7, 5767–5779. [Google Scholar] [CrossRef] [PubMed]
- Cheng, P.H.; Rao, X.M.; McMasters, K.M.; Zhou, H.S. Molecular basis for viral selective replication in cancer cells: activation of CDK2 by adenovirus-induced cyclin E. PLoS ONE 2013, 8, e57340. [Google Scholar] [CrossRef] [PubMed]
- Cheng, P.H.; Rao, X.M.; Duan, X.; Li, X.F.; Egger, M.E.; McMasters, K.M.; Zhou, H.S. Virotherapy targeting cyclin E overexpression in tumors with adenovirus-enhanced cancer-selective promoter. J. Mol. Med. 2015, 93, 211–223. [Google Scholar] [CrossRef] [PubMed]
- Debbas, M.; White, E. Wild-type p53 mediates apoptosis by E1A, which is inhibited by E1B. Genes Dev. 1993, 7, 546–554. [Google Scholar] [CrossRef] [PubMed]
- Rao, L.; Debbas, M.; Sabbatini, P.; Hockenbery, D.; Korsmeyer, S.; White, E. The adenovirus E1A proteins induce apoptosis, which is inhibited by the E1B 19-kDa and Bcl-2 proteins. Proc. Natl. Acad. Sci. USA 1992, 89, 7742–7746. [Google Scholar] [CrossRef] [PubMed]
- White, E.; Sabbatini, P.; Debbas, M.; Wold, W.S.; Kusher, D.I.; Gooding, L.R. The 19-kilodalton adenovirus E1B transforming protein inhibits programmed cell death and prevents cytolysis by tumor necrosis factor alpha. Mol. Cell. Biol. 1992, 12, 2570–2580. [Google Scholar] [CrossRef] [PubMed]
- Cuconati, A.; Degenhardt, K.; Sundararajan, R.; Anschel, A.; White, E. Bak and Bax function to limit adenovirus replication through apoptosis induction. J. Virol. 2002, 76, 4547–4558. [Google Scholar] [CrossRef] [PubMed]
- Bischoff, J.R.; Kim, D.H.; Williams, A.; Heise, C.; Horn, S.; Muna, M.; Ng, L.; Nye, J.A.; SampsonJohannes, A.; Fattaey, A.; et al. An adenovirus mutant that replicates selectively in p53-deficient human tumor cells. Science 1996, 274, 373–376. [Google Scholar] [CrossRef]
- Kirn, D. Oncolytic virotherapy for cancer with the adenovirus dl1520 (Onyx-015): results of phase I and II trials. Expert Opin. Biol. Ther. 2001, 1, 525–538. [Google Scholar] [CrossRef] [PubMed]
- Kirn, D. Clinical research results with dl1520 (Onyx-015), a replication-selective adenovirus for the treatment of cancer: What have we learned? Gene Ther. 2001, 8, 89–98. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Fang, H. Clinical trials with oncolytic adenovirus in China. Curr. Cancer Drug Targets. 2007, 7, 141–148. [Google Scholar] [CrossRef] [PubMed]
- Cafferata, E.G.; Maccio, D.R.; Lopez, M.V.; Viale, D.L.; Carbone, C.; Mazzolini, G.; Podhajcer, O.L. A novel A33 promoter-based conditionally replicative adenovirus suppresses tumor growth and eradicates hepatic metastases in human colon cancer models. Clin. Cancer Res. 2009, 15, 3037–3049. [Google Scholar] [CrossRef] [PubMed]
- Nemunaitis, J.; Tong, A.W.; Nemunaitis, M.; Senzer, N.; Phadke, A.P.; Bedell, C.; Adams, N.; Zhang, Y.A.; Maples, P.B.; Chen, S.; et al. A phase I study of telomerase-specific replication competent oncolytic adenovirus (telomelysin) for various solid tumors. Mol. Ther. 2010, 18, 429–434. [Google Scholar] [CrossRef] [PubMed]
- DeWeese, T.L.; van der Poel, H.; Li, S.; Mikhak, B.; Drew, R.; Goemann, M.; Hamper, U.; DeJong, R.; Detorie, N.; Rodriguez, R.; et al. A phase I trial of CV706, a replication-competent, PSA selective oncolytic adenovirus, for the treatment of locally recurrent prostate cancer following radiation therapy. Cancer Res. 2001, 61, 7464–7472. [Google Scholar] [PubMed]
- Cheng, P.H.; Lian, S.; Zhao, R.; Rao, X.M.; McMasters, K.M.; Zhou, H.S. Combination of autophagy inducer rapamycin and oncolytic adenovirus improves antitumor effect in cancer cells. Virol. J. 2013, 10, 293. [Google Scholar] [CrossRef] [PubMed]
- Malumbres, M.; Barbacid, M. To cycle or not to cycle: a critical decision in cancer. Nat. Rev. Cancer. 2001, 1, 222–231. [Google Scholar] [CrossRef] [PubMed]
- Cheng, P.H.; Rao, X.M.; Duan, X.; Li, X.F.; Egger, M.E.; McMasters, K.M.; Zhou, H.S. Virotherapy targeting cyclin E overexpression in tumors with adenovirus-enhanced cancer-selective promoter. J. Mol. Med. 2015, 93, 211–223. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, M.; Curiel, D.T. Current issues and future directions of oncolytic adenoviruses. Mol. Ther. 2010, 18, 243–250. [Google Scholar] [CrossRef] [PubMed]
- McCormick, F. Cancer-specific viruses and the development of ONYX-015. Cancer Biol Ther. 2003, 2, S157–S160. [Google Scholar] [CrossRef] [PubMed]
- Russell, S.J.; Peng, K.W.; Bell, J.C. Oncolytic virotherapy. Nat. Biotechnol. 2012, 30, 658–670. [Google Scholar] [CrossRef] [PubMed]
- Wechman, S.L.; Rao, X.M.; Cheng, P.H.; Gomez-Gutierrez, J.G.; McMasters, K.M.; Zhou, H.S. Development of an Oncolytic Adenovirus with Enhanced Spread Ability through Repeated UV Irradiation and Cancer Selection. Viruses 2016, 8, 6. [Google Scholar] [CrossRef] [PubMed]
- Jones, N.; Shenk, T. Isolation of adenovirus type 5 host range deletion mutants defective for transformation of rat embryo cells. Cell 1979, 17, 683–689. [Google Scholar] [CrossRef]
- Rao, X.M.; Tseng, M.T.; Zheng, X.; Dong, Y.; Jamshidi-Parsian, A.; Thompson, T.C.; Brenner, M.K.; McMasters, K.M.; Zhou, H.S. E1A-induced apoptosis does not prevent replication of adenoviruses with deletion of E1b in majority of infected cancer cells. Cancer Gene Ther. 2004, 11, 585–593. [Google Scholar] [CrossRef] [PubMed]
- Sandig, V.; Youil, R.; Bett, A.J.; Franlin, L.L.; Oshima, M.; Maione, D.; Wang, F.; Metzker, M.L.; Savino, R.; Caskey, C.T. Optimization of the helper-dependent adenovirus system for production and potency in vivo. Proc. Natl. Acad. Sci. USA 2000, 97, 1002–1007. [Google Scholar] [CrossRef] [PubMed]
- Ishiyama, M.; Tominaga, H.; Shiga, M.; Sasamoto, K.; Ohkura, Y.; Ueno, K. A combined assay of cell viability and in vitro cytotoxicity with a highly water-soluble tetrazolium salt, neutral red and crystal violet. Biol. Pharm. Bull. 1996, 19, 1518–1520. [Google Scholar] [CrossRef] [PubMed]
- Graham, F.L.; Rudy, J.; Brinkley, P. Infectious Circular DNA of Human Adenovirus Type-5 - Regeneration of Viral-DNA Termini from Molecules Lacking Terminal Sequences. EMBO J. 1989, 8, 2077–2085. [Google Scholar] [PubMed]
- Zhou, H.S.; ONeal, W.; Morral, N.; Beaudet, A.L. Development of a complementing cell line and a system for construction of adenovirus vectors with E1 and E2a deleted. J. Virol. 1996, 70, 7030–7038. [Google Scholar] [PubMed]
- Bett, A.J.; Haddara, W.; Prevec, L.; Graham, F.L. An efficient and flexible system for construction of adenovirus vectors with insertions or deletions in early regions 1 and 3. Proc. Natl. Acad. Sci. USA 1994, 91, 8802–8806. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Low, J.A.; Christensen, J.B.; Imperiale, M.J. Role for the adenovirus IVa2 protein in packaging of viral DNA. J. Virol. 2001, 75, 10446–10454. [Google Scholar] [CrossRef] [PubMed]
- Pardo-Mateos, A.; Young, C.S. Adenovirus IVa2 protein plays an important role in transcription from the major late promoter in vivo. Virology 2004, 327, 50–59. [Google Scholar] [CrossRef] [PubMed]
- Webster, A.; Leith, I.R.; Hay, R.T. Domain organization of the adenovirus preterminal protein. J. Virol. 1997, 71, 539–547. [Google Scholar] [PubMed]
- Webster, A.; Leith, I.R.; Nicholson, J.; Hounsell, J.; Hay, R.T. Role of preterminal protein processing in adenovirus replication. J. Virol. 1997, 71, 6381–6389. [Google Scholar] [PubMed]
- Tamanoi, F.; Stillman, B.W. Function of adenovirus terminal protein in the initiation of DNA replication. Proc. Natl. Acad. Sci. USA 1982, 79, 2221–2225. [Google Scholar] [CrossRef] [PubMed]
- Van Breukelen, B.; Brenkman, A.B.; Holthuizen, P.E.; van der Vliet, P.C. Adenovirus type 5 DNA binding protein stimulates binding of DNA polymerase to the replication origin. J. Virol. 2003, 77, 915–922. [Google Scholar] [CrossRef] [PubMed]
- Gustin, K.E.; Imperiale, M.J. Encapsidation of viral DNA requires the adenovirus L1 52/55-kilodalton protein. J. Virol. 1998, 72, 7860–7870. [Google Scholar] [PubMed]
- Hasson, T.B.; Ornelles, D.A.; Shenk, T. Adenovirus L1 52- and 55-kilodalton proteins are present within assembling virions and colocalize with nuclear structures distinct from replication centers. J. Virol. 1992, 66, 6133–6142. [Google Scholar] [PubMed]
- Paterson, C.P.; Ayalew, L.E.; Tikoo, S.K. Mapping of nuclear import signal and importin alpha3 binding regions of 52K protein of bovine adenovirus-3. Virology 2012, 432, 63–72. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, P.K.; Vayda, M.E.; Flint, S.J. Identification of proteins and protein domains that contact DNA within adenovirus nucleoprotein cores by ultraviolet light crosslinking of oligonucleotides 32P-labelled in vivo. J. Mol. Biol. 1986, 188, 23–37. [Google Scholar] [CrossRef]
- Chatterjee, P.K.; Vayda, M.E.; Flint, S.J. Interactions among the three adenovirus core proteins. J. Virol. 1985, 55, 379–386. [Google Scholar] [PubMed]
- Matthews, D.A.; Russell, W.C. Adenovirus core protein V interacts with p32--a protein which is associated with both the mitochondria and the nucleus. J. Gen. Virol. 1998, 79, 1677–1685. [Google Scholar] [CrossRef] [PubMed]
- Alva, V.; Ammelburg, M.; Soding, J.; Lupas, A.N. On the origin of the histone fold. BMC Struct. Biol. 2007, 7, 17. [Google Scholar] [CrossRef] [PubMed]
- Perez-Vargas, J.; Vaughan, R.C.; Houser, C.; Hastie, K.M.; Kao, C.C.; Nemerow, G.R. Isolation and characterization of the DNA and protein binding activities of adenovirus core protein V. J. Virol. 2014, 88, 9287–9296. [Google Scholar] [CrossRef] [PubMed]
- Xue, B.; Dunbrack, R.L.; Williams, R.W.; Dunker, A.K.; Uversky, V.N. PONDR-FIT: A meta-predictor of intrinsically disordered amino acids. Biochim. Biophys. Acta 2010, 1804, 996–1010. [Google Scholar] [CrossRef] [PubMed]
- Tormanen, H.; Backstrom, E.; Carlsson, A.; Akusjarvi, G. L4–33K, an adenovirus-encoded alternative RNA splicing factor. J. Biol. Chem. 2006, 281, 36510–36517. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.; Guimet, D.; Hearing, P. The adenovirus L4–33K protein regulates both late gene expression patterns and viral DNA packaging. J. Virol. 2013, 87, 6739–6747. [Google Scholar] [CrossRef] [PubMed]
- Ali, H.; LeRoy, G.; Bridge, G.; Flint, S.J. The adenovirus L4 33-kilodalton protein binds to intragenic sequences of the major late promoter required for late phase-specific stimulation of transcription. J. Virol. 2007, 81, 1327–1338. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.C.; Hearing, P. Adenovirus structural protein IIIa is involved in the serotype specificity of viral DNA packaging. J. Virol. 2011, 85, 7849–7855. [Google Scholar] [CrossRef] [PubMed]
- Shenk, T.J. Adenoviridae: The Viruses and Their Replication. In Fields Virology; Knipe, D.M., Howley, P.M., Eds.; Lippincott-Raven: Philadelphia, PA, USA, 1996; pp. 2111–2148. [Google Scholar]
- Bergstrom Lind, S.; Artemenko, K.A.; Elfineh, L.; Zhao, Y.; Bergquist, J.; Pettersson, U. Post translational modifications in adenovirus type 2. Virology 2013, 447, 104–111. [Google Scholar] [CrossRef] [PubMed]
- Mangel, W.F.; San Martin, C. Structure, function and dynamics in adenovirus maturation. Viruses 2014, 6, 4536–4570. [Google Scholar] [CrossRef] [PubMed]
- Vellinga, J.; Van der Heijdt, S.; Hoeben, R.C. The adenovirus capsid: major progress in minor proteins. J. Gen. Virol. 2005, 86, 1581–1588. [Google Scholar] [CrossRef] [PubMed]
- Molinier-Frenkel, V.; Lengagne, R.; Gaden, F.; Hong, S.S.; Choppin, J.; Gahery-Segard, H.; Boulanger, P.; Guillet, J.G. Adenovirus hexon protein is a potent adjuvant for activation of a cellular immune response. J. Virol. 2002, 76, 127–135. [Google Scholar] [CrossRef] [PubMed]
- Bruder, J.T.; Semenova, E.; Chen, P.; Limbach, K.; Patterson, N.B.; Stefaniak, M.E.; Konovalova, S.; Thomas, C.; Hamilton, M.; King, C.R.; et al. Modification of Ad5 hexon hypervariable regions circumvents pre-existing Ad5 neutralizing antibodies and induces protective immune responses. PLoS ONE 2012, 7, e33920. [Google Scholar] [CrossRef]
- Arnold, U.; Raines, R.T. Replacing a single atom accelerates the folding of a protein and increases its thermostability. Org. Biomol. Chem. 2016, 14, 6780–6785. [Google Scholar] [CrossRef] [PubMed]
- Reddy, V.S.; Nemerow, G.R. Structures and organization of adenovirus cement proteins provide insights into the role of capsid maturation in virus entry and infection. Proc. Natl. Acad. Sci. USA 2014, 111, 11715–11720. [Google Scholar] [CrossRef] [PubMed]
- Reddy, V.S.; Natchiar, S.K.; Stewart, P.L.; Nemerow, G.R. Crystal structure of human adenovirus at 3.5 A resolution. Science 2010, 329, 1071–1075. [Google Scholar] [CrossRef] [PubMed]
- Nettelbeck, D.M.; Jerome, V.; Muller, R. Gene therapy: designer promoters for tumour targeting. Trends Genet. 2000, 16, 174–181. [Google Scholar] [CrossRef]
- Yan, W.; Kitzes, G.; Dormishian, F.; Hawkins, L.; Sampson-Johannes, A.; Watanabe, J.; Holt, J.; Lee, V.; Dubensky, T.; Fattaey, A.; et al. Developing novel oncolytic adenoviruses through bioselection. J. Virol. 2003, 77, 2640–2650. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Zhu, Z.; Tang, L.; Wang, L.; Tan, X.; Yu, P.; Zhang, Y.; Tian, X.; Wang, J.; Zhang, Y.; et al. Genomic analyses of recombinant adenovirus type 11a in China. J. Clin. Microbiol. 2009, 47, 3082–3090. [Google Scholar] [CrossRef] [PubMed]
- Huebner, R.J.; Rowe, W.P.; Schatten, W.E.; Smith, R.R.; Thomas, L.B. Studies on the use of viruses in the treatment of carcinoma of the cervix. Cancer 1956, 9, 1211–1218. [Google Scholar] [PubMed]
- Klein, S.R.; Piya, S.; Lu, Z.; Xia, Y.; Alonso, M.M.; White, E.J.; Wei, J.; Gomez-Manzano, C.; Jiang, H.; Fueyo, J. C-Jun N-terminal kinases are required for oncolytic adenovirus-mediated autophagy. Oncogene 2015, 34, 5295–5301. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Gomez-Manzano, C.; Alemany, R.; Medrano, D.; Alonso, M.; Bekele, B.N.; Lin, E.; Conrad, C.C.; Yung, W.K.; Fueyo, J. Comparative effect of oncolytic adenoviruses with E1A-55 kDa or E1B-55 kDa deletions in malignant gliomas. Neoplasia 2005, 7, 48–56. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Rocha, H.; Gomez-Gutierrez, J.G.; Garcia-Garcia, A.; Rao, X.M.; Chen, L.; McMasters, K.M.; Zhou, H.S. Adenoviruses induce autophagy to promote virus replication and oncolysis. Virology 2011, 416, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Baehrecke, E.H. Autophagy: dual roles in life and death? Nat. Rev. Mol. Cell Biol. 2005, 6, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Klionsky, D.J.; Abeliovich, H.; Agostinis, P.; Agrawal, D.K.; Aliev, G.; Askew, D.S.; Baba, M.; Baehrecke, E.H.; Bahr, B.A.; Ballabio, A.; et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy 2008, 4, 151–175. [Google Scholar] [CrossRef] [PubMed]
- Klionsky, D.J.; Emr, S.D. Autophagy as a regulated pathway of cellular degradation. Science 2000, 290, 1717–1721. [Google Scholar] [CrossRef] [PubMed]
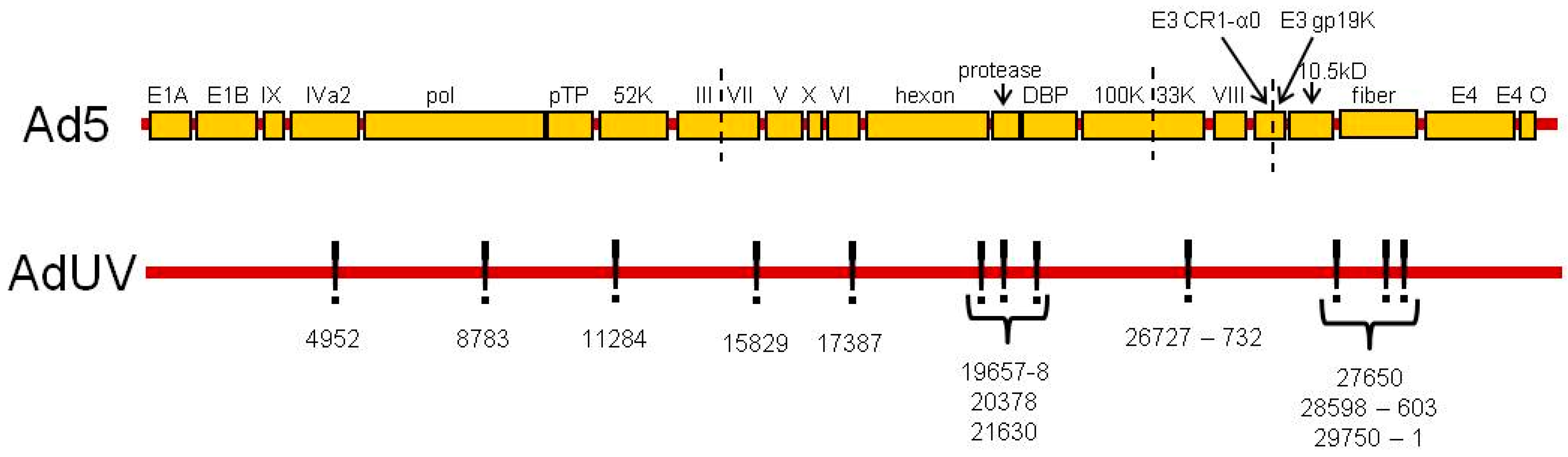
| Site | Gene | Mutation | aa Changed? | Non-Conservative Mutation? |
|---|---|---|---|---|
| 4952 | IVa2 | G to C | T to S | No, T and S are both polar uncharged aa |
| 8783 | pTP | G to A | R to L | Yes, R is basic (+) and L is hydrophobic |
| 11284 | 52K | T to C | Y to H | Yes, Y is hydrophobic and H is a positivity charged aa |
| 12728 | Intron | SN deletion | No | No |
| 15829 | pIII | SN deletion, non-sense mutation | G to A and lost 13 aa: IVSPRVLSSRTF | Yes, lost 13 aa |
| 16588 | pV | G to A | No | No |
| 17387 | pV | G to C | G to R | Yes, G is hydrophobic and R is basic (+) |
| 18755 | pVI | SN deletion | No, targeted only the last codon. Did not remove the codon stop | No |
| 19483 | Hexon | T to A | No | No |
| 19513 | Hexon | T to A | No | No |
| 19657–8 | Hexon | DN substitution, GA to AG | AT to AA | Yes, T is polar uncharged and A is hydrophobic |
| 20378 | Hexon | T to C | L to V | No, both L and V are hydrophobic |
| 21163 | Hexon | C to T | No | No |
| 21630 | Hexon | G to A | R to Q | Yes, R is basic (+) and Q is polar uncharged |
| 25995 | 100K | A to T | No | No |
| 26561 | Intron | G to A | No | No |
| 26727–32 | 33K | Six-nucleotide deletion | Lost 2 aa A and A | Yes, lost 2 aa |
| 27161 | Intron | C to T | No | No |
| 27314 | pVIII | C to A | No | No |
| 27339 | pVIII | T to C | No | No |
| 27650–1 | pVIII | DN substitution TC to CT | RP to RS | Yes, P is cyclic and S is polar uncharged |
| 28120 | Intron | T to C | No | No |
| 28596–601 | E3 CR1-α0 | Six-nucleotide deletion | Lost 2 aa I and G | Yes, lost 2 aa |
| 29750 | E3A 10.5kD | T to G | M to R | Yes, M is hydrophobic and R is basic (+) |
| 35776 | Intron | A to C | No | No |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wechman, S.L.; Rao, X.-M.; McMasters, K.M.; Zhou, H.S. Adenovirus with DNA Packaging Gene Mutations Increased Virus Release. Viruses 2016, 8, 333. https://doi.org/10.3390/v8120333
Wechman SL, Rao X-M, McMasters KM, Zhou HS. Adenovirus with DNA Packaging Gene Mutations Increased Virus Release. Viruses. 2016; 8(12):333. https://doi.org/10.3390/v8120333
Chicago/Turabian StyleWechman, Stephen L., Xiao-Mei Rao, Kelly M. McMasters, and Heshan Sam Zhou. 2016. "Adenovirus with DNA Packaging Gene Mutations Increased Virus Release" Viruses 8, no. 12: 333. https://doi.org/10.3390/v8120333
APA StyleWechman, S. L., Rao, X.-M., McMasters, K. M., & Zhou, H. S. (2016). Adenovirus with DNA Packaging Gene Mutations Increased Virus Release. Viruses, 8(12), 333. https://doi.org/10.3390/v8120333





